Haemophilus influenzae can cause a variety of infections, including otitis media, bronchitis, pneumonia and meningitis [Reference Murphy and Apicella1, Reference Saito2]. In the past, the activity of two β-lactamases, TEM-1 and ROB-1, accounted for almost all isolates with decreased susceptibility to ampicillin [Reference Felmingham and Washington3]. At present, the global prevalence of β-lactamase-negative ampicillin-resistant (BLNAR) H. influenzae remains low [Reference Karlowsky4, Reference Hoban and Felmingham5], but the proportion of clinical BLNAR isolates is rapidly increasing, and has now reached more than 20% in Japan [Reference Suzuki, Nishimura and Baba6]. The characteristics of antimicrobial resistance of these strains are a serious concern for clinical prescribing. BLNAR strains have a resistance mechanism that decreases the affinity of ampicillin for penicillin-binding proteins (PBPs) [Reference Ubukata7]. The resistance phenotypes are classified according to substitutions at three positions of the ftsI gene which mediates septal peptidoglycan sysnthesis allowing the classification of strains as BLNAR or low-BLNAR by PCR [Reference Hasegawa8].
In total, 163 strains of H. influenzae were isolated from sputum from patients with respiratory tract infections in Nagasaki University and its affiliated hospitals. These strains were selected at random and were divided into four groups, 45 strains between 1987 and 1989, 32 between 1991 and 1993, 41 between 1995 and 1997, and 45 between 1998 and 2000. Strains were capsule typed by slide agglutination with antisera (Difco Laboratories, Detroit, MI, USA) and β-lactamase production was detected using a nitrocefin-impregnated disk (Becton Dickinson, Sparks, MD, USA). The minimum inhibitory concentration (MIC) of five antibiotics was determined by the agar dilution method according to the guidelines of the National Committee for Clinical Laboratory Standards [9]. The antibiotics were: ampicillin (Meiji Seika Kaisha, Tokyo, Japan), levofloxacin (Daiichi Pharmaceutical Co., Tokyo), cefditoren (Meiji Seika Kaisha), cefdinir (Astellas Pharma Inc., Tokyo) and ceftriaxone (Chugai Pharmaceutical Co., Tokyo).
PCR was performed to identify resistance genes using a multiplex assay as described previously [Reference Hasegawa8]. Four sets of primers were obtained from Wakunaga Pharmaceutical Co. (Hiroshima, Japan): P6 primers to amplify the P6 gene which encodes the P6 membrane protein specific for H. influenzae; TEM-1 primers to amplify a part of the bla TEM-1 gene; PBP3-S primers to identify an Asn526→Lys amino-acid substitution in the ftsI gene; and PBP3-BLN primers to identify an Asn526→Lys and Ser385→Thr amino-acid substitution in the ftsI gene.
Pulsed-field gel electrophoresis (PFGE) was performed as described previously [Reference Yano10] using SmaI digestion (Takara Shuzo Co., Shiga, Japan) and electrophoresis in a CHEF Mapper PFGE system (Bio-Rad Life Science Group, Hercules, CA, USA) was carried out at 6 V/cm with switch times of 0·47 and 63 s, and a run-time of 20 h. After staining with ethidium bromide, the interpretation of PFGE patterns was based on the criteria described by Tenover et al. [Reference Tenover11]. Briefly, PFGE patterns were classified into four groups: identical in profile=indistinguishable; 1–3 bands difference=closely related; 4–6 bands difference=possibly related; and >7 bands difference=different.
The Table shows that of the 45 strains isolated from 1987 to 1989, 37 (82·2%) were classified as β-lactamase-negative ampicillin-susceptible (BLNAS) strains, seven strains produced TEM-1-type β-lactamase and were ampicillin resistant (BLPAR) and one strain was classified as low-BLNAR by PCR. The proportion of BLNAS strains fell in the ensuing two sampling periods but recovered in the final time period to 80%. The frequency of BLPAR strains fluctuated from 15·6% in the initial period through 6·3% and 12·2% to 2·2% in the final sampling period. Similar variation was observed for low-BLNAR strains with just 2·2% of strains expressing this phenotype in the first period but rising to almost 20% in the third period before falling back to 11%. Three BLNAR strains (6·7%) were detected only in the fourth sampling period. The respective MIC80 values (μg/ml) for the four periods against the strain collection is also shown in the Table. The NCCLS susceptibility/resistant break-points for H. influenzae are 1 μg/ml for ampicillin, 2 μg/ml for levofloxacin, 1 μg/ml for cefdinir, and 2 μg/ml for ceftriaxone. The MIC of ampicillin, ceftriaxone, cefditoren and levofloxacin remained constant or within one doubling concentration over the years but resistance to cefdinir increased by eight-fold over the sampling period (Table).
Table. Annual changes of the prevalence of each resistance class and antimicrobial susceptibility to five antibiotics
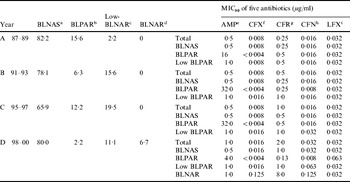
a β-lactamase-negative ampicillin susceptible strains.
b β-lactamase-producing ampicillin-resistant (TEM-1 type) strains.
c Low β-lactamase-negative ampicillin-resistant strains.
d β-lactamase-negative ampicillin-resistant strains.
e Ampicillin.
f Ceftriaxone.
g Cefdinir.
h Cefditoren.
i Levofloxacin.
The three BLNAR strains gave distinct DNA profiles in PFGE while 11 profiles were distinguished among the 17 low-BLNAR strains. Two of these profiles exhibited some similarities to profiles found in fully BLNAR strains. Three pairs of strains each exhibited similar patterns and four strains were grouped within the same pattern (Fig.). All but two of the 20 strains were non-typable with capsular antisera.

Fig. PFGE patterns of SmaI-digested DNA from three BLNAR and 17 low-BLNAR H. influenzae strains. Lanes 1–3, BLNAR strains; lanes 4–20, low-BLNAR strains. Coding of the PFGE patterns depicting group and subgroups and serotype status. N, Non-typable.
H. influenzae is one of the important pathogens associated with respiratory tract infections and thus acquisition of antimicrobial resistance raises concern. The prevalence of BLNAR strains was reported to be 2·4% in the United States between 2002 and 2003 [Reference Heilmann12], 1·3% in France in 1999 [Reference Dabernat13], and 9·3% in Spain between 1998 and 1999 [Reference Marco14]. Nevertheless, their global prevalence remains relatively low. However, BLNAR strains are spreading rapidly with increasing frequency in Japan with reported prevalence rates of 14·9% between 1996 and 1997 [Reference Seki15], and 23·1% between 1998 and 1999 [Reference Suzuki, Nishimura and Baba6], although BLNAR was identified by only MIC in these reports. We report here a prevalence of BLNAR strains of 6·7% by PCR between 1998 and 2000. An understanding of the characteristics of antimicrobial resistance of H. influenzae, especially BLNAR strains, is important not only for prescribing clinicians but also for formulation of practicable chemotherapy guidelines. Although H. influenzae strains are generally susceptible to the early cephalosporins [Reference Hoban and Felmingham5, Reference Heilmann12], the BLNAR and low-BLNAR strains recovered here showed a marginal increase in MIC to two of the three cephalosporins tested and an eight-fold increase in MIC of cefdinir which is consistent with a previous report from Japan [Reference Suzuki, Nishimura and Baba6].
PFGE of DNA macrorestriction fragments is a sensitive fingerprinting method for H. influenzae and this method was used by Karlowsky et al. [Reference Karlowsky4] to demonstrate clonal dissemination of BLNAR strains in the United States between 2000 and 2001. However, the BLNAR and low-BLNAR strains found here displayed a variety of genetic backgrounds. It has previously been reported that H. influenzae, including resistant strains, can be transmitted at day-care centres or in the home [Reference Dabernat13, Reference Watanabe16], and this may be one reason for the spread of BLNAR strains in Japan. We did observe that some low-BLNAR strains isolated from different patients had similar PFGE patterns and therefore must consider that such strains could potentially spread in a community. Ongoing monitoring of H. influenzae resistance determinants is thought to be important and may help to predict how this organism responds to current antimicrobial regimens [Reference Rennie and Ibrahim17]. Despite the limitations of this pilot study, particularly the small sample size, it shows the value of surveillance of antimicrobial resistance levels and their genetic determinants in H. influenzae and further surveys specifically of BLNAR strains in the wider Japanese community should be undertaken to inform antimicrobial prescribing policy.
ACKNOWLEDGEMENTS
We thank Akihiro Wada (Department of Bacteriology, Institute of Tropical Medicine, Nagasaki University, Japan), Chieko Shimauchi (Miyazaki Prefectural Nursing University, Japan) and Matsuhisa Inoue (Kitasato University School of Medicine, Japan) for their help in completing the PFGE studies. The study was supported by U.S.–Japan Cooperative Medical Science Program, Acute Respiratory Infections Panel.
DECLARATION OF INTEREST
None.






