INTRODUCTION
Shigella spp. remain a major public health problem in both developing and developed countries [Reference Ram1]. They are estimated to be responsible for over 146 million global infections and about 1·1 million people die from shigellosis each year [Reference Kotloff2]. The majority (61%) of deaths occur in children aged <5 years [Reference Kotloff2]. The genus Shigella is composed of four species, Shigella dysenteriae, S. flexneri, S. boydii, and S. sonnei. S. sonnei is the most commonly reported Shigella spp. in industrialized countries and it is often the second most prevalent Shigella spp. in developing countries after S. flexneri [Reference Kotloff2, Reference Niyogi3]. Except for S. sonnei, each species contains multiple serotypes, based on the structure of the O antigen [Reference Simmons and Romanowska4]. Extensive studies with respect to S. sonnei's clonal diversity and antimicrobial resistance pattern have not been undertaken in the developing world [Reference Talukder5].
There is little epidemiological information available regarding the burden of shigellosis in Egypt; however, a prospective diarrhoea surveillance study performed in two villages in the Nile Delta region of Egypt during 1995–1998 indicated that the incidence of Shigella in children with diarrhoea aged <3 years was 0·19 episodes per child-year [Reference Abu-Elyazeed6]. Incidence was shown to vary from year to year with the highest incidence observed in children aged between 12 and 23 months. S. flexneri was the dominant pathogen (55%) followed distantly by S. sonnei (22%), S. dysenteriae (19%), and S. boydii (2%). In a separate and unlinked study, Weirzba et al. [Reference Wierzba7] examined children aged ⩽5 years presenting at two medical facilities in the Nile Delta from 2000 to 2002. In this study, 2% of diarrhoeal stools were positive for Shigella (n = 23) by culture; 83% were S. flexneri while only 4% were S. sonnei. Although the objectives and populations under surveillance described by these two studies are not directly comparable, it is clear that Shigella spp. are a significant cause of diarrhoea for children in Egypt and S. flexneri is the prevailing species.
Shigella spp. have the ability to invade the epithelial cells of the intestinal mucosa with further dissemination to other cells using proteins encoded by genes located on an invasion plasmid [Reference Sansonetti, Kopecko and Formal8]. This 140 MDa invasion plasmid carries several genes encoding virulence factors such as the sen gene which encodes the Shigella enterotoxin 2 (ShET-2) [Reference Nataro9] and ipaH, encoding the invasion plasmid antigen IpaH, found in multiple copies on the invasion plasmid and on the chromosome [Reference Hale10].
Antibiotic therapy can limit the duration of shigellosis and shedding of the organism. However, resistance has been increasing to the most frequently administered antibiotics: trimethoprim–sulfamethoxazole, ampicillin and chloramphenicol [Reference Ahmed11–Reference Vlieghe13]. Quinolones and fluoroquinolones are effective in controlling the bacillus, although resistant isolates have been described [Reference MoezArdalan14]. Several mechanisms involving mobile genetic elements such as plasmids, integrons, and transposons contribute to the spread of antibiotic resistance in bacterial species [Reference Gassama-Sow15] and mobile genetic elements are frequently reported in Shigella isolates [Reference Mammina16–Reference Gassama-Sow18]. Unlike most enteric pathogens, S. sonnei is not known to have a natural reservoir other than humans, therefore antimicrobial resistance in S. sonnei may be expected to predominantly reflect gene transfer and selective pressures in the human gastrointestinal tract [Reference Zhang19, Reference Yang20]. S. sonnei isolates have a high prevalence of class 2 integrons; these integrons harbour dfrA1, sat and aadA1 cassettes, conferring resistance to trimethoprim/streptothricin and spectinomycin/streptomycin, respectively [Reference Gassama-Sow15, Reference Madiyarov21, Reference Zhu22].
Several phenotypic and molecular techniques have been used to type S. sonnei. Pulsed-field gel electrophoresis (PFGE), a broadly applicable typing method with a high degree of intra- and inter-laboratory reproducibility, has been previously applied in studies of this organism in several countries and allowed the identification of epidemiologically relevant strains [Reference Mammina23, Reference Jin24]. Other approaches used to characterize S. sonnei isolates include enterobacterial repetitive intergenic consensus PCR [Reference Penatti25], repetitive extragenic palindromic PCR [Reference Penatti25, Reference Navia26], inter-IS1 spacer typing [Reference Wei27] and multilocus variable-nucleotide tandem repeat analysis (MLVA) [Reference Liang28]. Recent reports suggest that PFGE and MLVA methods generate concordant phylogenies when applied to a global set of S. sonnei isolates [Reference Chiou29], although MLVA provides better discrimination for outbreak analyses [Reference Liang28].
In this study, we compared S. sonnei isolated from children seeking medical care (MC) and those enrolled in a prospective diarrhoea birth cohort (BC) study from the same regions in Egypt using both conventional and advanced molecular techniques. Collection and analysis of this information is critical for epidemiological investigations, and can be used to observe changes in circulating strains over time.
MATERIALS AND METHODS
Bacterial isolates
A total of 99 S. sonnei isolates were obtained from 1999 to 2005 as part of paediatric diarrhoeal surveillance projects based in two physically separate areas of Egypt. The isolates were obtained from two different studies. The first study (children seeking medical care for moderate to severe diarrhoea, MC) was conducted in Abu Homos District, Beheira Governorate, Nile Delta and in Mokattam Hills, Cairo. Children aged <5 years who attended outpatient clinics because of symptoms associated with diarrhoea were eligible for enrolment. A stool sample and two rectal swabs were collected and sent to the laboratory at NAMRU-3, Cairo. The second study (community-based cohort study, BC) consisted of four cohorts living in the Abu Homos District. Children were enrolled as soon after birth as possible, but before reaching the age of 1 year and were followed until reaching the second of third year of life, depending on the cohort. Social workers visited the children's homes twice per week and if the child had diarrhoea, a stool specimen and two rectal swabs were collected and sent to the laboratory at NAMRU-3. As part of all these studies, routine bacterial culture was performed for Aeromonas, Campylobacter, enterotoxigenic Escherichia coli (ETEC), and Salmonella. We were able to identify S. sonnei isolates from 51 children seeking medical care in the MC study and 48 children enrolled in the BC study; the majority of BC children had diarrhoea (n = 41). Isolates were identified as S. sonnei using standard microbiological and biochemical procedures. Confirmation was made using both API-20E biochemical test strips (bioMérieux SA, France) and slide agglutination test using Shigella group antisera (Becton Dickenson, USA) according to the manufacturer's instructions. Biotyping of S. sonnei isolates was performed using standard methods for fermentation of rhamnose and xylose and hydrolysis of ortho- nitrophenyl-β-d-galactopyranoside (ONPG) [Reference Mammina16].
Antimicrobial susceptibility testing
Isolates were tested for their susceptibility to ampicillin (AMP, 10 μg), cephalothin (CR, 30 μg), streptomycin (STR, 10 μg), chloramphenicol (CHL, 30 μg), nalidixic acid (NA, 30 μg), tetracycline (TET, 30 μg), trimethoprim/sulfamethoxazole (SXT, 25 μg) and ciprofloxacin (CIP, 5 μg) by the disk diffusion method according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) [30]. Antimicrobial disks were obtained from Becton Dickinson. E. coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were used as quality control organisms.
Plasmid analysis
Plasmid DNA was extracted from an overnight bacterial culture on Mueller–Hinton agar (Hardy Diagnostics, USA) supplemented with 5% sheep blood using the JETstar Plasmid Purification System (Genomed GmbH, Germany). The extracted plasmid DNA was separated by electrophoresis in a 0·8% agarose gel using 1× Tris-acetate-EDTA buffer at 120 V for 1 h.
PCR assays
S. sonnei isolates were evaluated for the presence of ipaH, set, sen, class I and class II integrase genes by PCR. Whole-cell lysates were prepared using a 10 μl loop full of fresh overnight culture on MacConkey agar (Oxoid, UK) suspended in 400 μl sterile deionized water as previously described [Reference Houng, Sethabutr and Echeverria31]. Amplification of the targeted genes was performed using: ipaH-F (5′-GTT CCT TGA CCG CCT TTC CGA TAC CGT C-3′) and ipaH-R (5′-GCC GGT CAG CCA CCC TCT GAG AGT AC-3′) [Reference Sethabutr32]; set-F (5′-GTG AAC CTG CTG CCG ATA TC-3′) and set-R (5′-ATT TGT GGA TAA AAA TGA CG-3′) [Reference Vargas33]; sen-F (5′-CGT TAG AAT TAC TTT TGG CAG C-3′) and sen-R (5′-GGC CAG CAA ATT TAC AAT ATC C-3′) [Reference Yavzori, Cohen and Orr34]; int1F (5′-CCT CCC GCA CGA TGA TC-3′) and intI1R (5′-TCC ACG CAT CGT CAG GC-3′), and intI2F (5′-TTA TTG CTG GGA TTA GGC-3′) and intI2R (5′-ACG GCT ACC CTC TGT TAT C-3′) [Reference Goldstein35]. PCR was conducted in a 25-μl reaction mixture containing 10 mm Tris–HCl (pH 8·3), 50 mm KCl, 1·5 mm MgCl2, 0·2 mm each deoxynucleoside triphosphate, 1·25 U of AmpliTaq DNA polymerase (Applied Biosystems, USA) using 2 μl bacterial cell lysate. All PCR amplifications were performed using the following amplification scheme: one cycle of denaturation at 95°C for 1 min, annealing at 52°C for 1 min, and extension at 72°C for 1 min, with a final extension at 72°C for 7 min. The reactions were performed in a GeneAmp PCR System 9700 (Applied Biosystems) and analysed by gel electrophoresis on 2·0% agarose (Sigma-Aldrich, Germany) gels using 1× Tris-acetate-EDTA buffer.
PFGE
Preparation of genomic DNA from S. sonnei isolates and separation of macrorestriction enzyme fragments were performed as described previously [Reference Ribot36]. Embedded genomic DNA was digested with 20 U of XbaI (New England Biolabs, USA) for 18 h at 37°C and DNA fragments were separated in 1% agarose gel suspended in 0·5× Tris-borate-EDTA buffer using a contour-clamped homogeneous electric field apparatus (CHEF-DRIII, Bio-Rad Laboratories, USA). Salmonella enterica serovar Braenderup strain H9812 restricted with XbaI was used as molecular weight standard as recommended previously [Reference Ribot36]. Software-assisted analysis of the PFGE banding patterns was performed using Bionumerics software (version 5.10; Applied Maths, USA). Genetic similarities between isolates were inferred from dendrograms created using the complete linkage method analysis of the XbaI macrorestriction PFGE patterns using the Dice coefficient with a 1·2% tolerance for the band migration distance [Reference DeLappe37]; this analysis method is considered a more strict interpretation of banding patterns than UPGMA [Reference DeLappe38].
Statistical analysis
Demographic and clinical data were described and compared across the two studies. Statistical comparisons were made using the χ2 test for comparison among proportions, Fisher's exact test or the Kruskal–Wallis test as appropriate. Analysis was conducted using SPSS version 17 (SPSS Inc., USA), and statistical significance was set at P = 0·05.
RESULTS
Epidemiological characteristics
S. sonnei were isolated from 99 children who were enrolled in BC (48, 48·5%) or MC (51, 51·5%) studies. Children enrolled in the MC study had a significantly longer duration of illness (P<0·018) coupled with more severe disease, judged by vomiting (P<0·001) and fever (P<0·0001), than children enrolled in the BC study (Table 1). Mild to moderate dehydration, however, was significantly higher (P<0·0001) in BC children. ETEC was the most common co-pathogen associated with S. sonnei infection (n = 17, 17·2%). Co-infection was more common in children enrolled in the MC study (22·9%), compared to children enrolled in the BC study (11·8%); however, this difference was not statistically significant. Rotavirus was the second most common pathogen associated with S. sonnei infection (n = 9, 9·1%), and was also more common in MC children (12·6% vs. 5·9%), although the difference was not statistically significant. We performed a separate analysis where cases with co-pathogens were removed (n = 17, Table 2) and found that MC children had statistically significant increases in diarrhoeal duration, vomiting, and fever compared to BC children either in the presence or absence of a detected co-pathogen (P = 0·018 or 0·032 for diarrhoeal duration, P = 0·001 or 0·01 for vomiting, P<0·0001 or 0·0001 for fever, respectively). Similarly, BC children were more likely to be dehydrated than MC children either in the presence or absence of a detected co-pathogen (P<0·0001 or 0·0001, respectively).
Table 1. Clinical and microbiological characteristics of Shigella sonnei culture-positive paediatric cases

IQR, Interquartile range; ETEC, enterotoxigenic Escherichia coli.
* Children seeking medical care for diarrhoea-associated symptoms.
† Children enrolled in a community-based diarrhoeal birth cohort study.
‡ Statistically significant.
§ 98 cases were assessed for blood in stool.
|| 94 cases were assessed for dehydration.
Table 2. Clinical and microbiological characteristics of Shigella sonnei culture-positive paediatric cases where S. sonnei was the sole pathogen detected

IQR, Interquartile range.
* Children seeking medical care for diarrhoea-associated symptoms.
† Children enrolled in a community-based diarrhoeal birth cohort study.
‡ Statistically significant.
§ 69 cases were assessed for dehydration.
Phenotypical characteristics
Three different phenotypes were recognized by biochemical testing using API-20E. Almost all (n = 97) of the isolates produced β-galactosidase, ornithine decarboxylase and fermented glucose, mannitol and arabinose. Two isolates (one each from the MC and BC studies) exhibited slightly different biochemical profiles. One of the isolates was unable to produce ornithine decarboxylase and the other fermented melibiose. Since all of the isolates produced β-galactosidase and could not ferment rhamnose or xylose, they were categorized as S. sonnei biotype g.
Multidrug resistance is common in Egyptian S. sonnei isolates
All 99 S. sonnei isolates were tested for susceptibility to a panel of eight antimicrobial agents (Table 3). We observed 100%, 98·9% and 92·9% resistance to STR, SXT, and TET, respectively. Greater variability was observed with both CR (50·5%) and AMP (12·1%) and resistance to CHL and NA was rare (1%). We did not detect any CIP-resistant isolates. Multiple resistances to TET/STR/SXT or TET/STR/SXT/CR were observed in 87 (87·8%) and 43 (43·4%) of all isolates, respectively. With the exception of AMP, no major differences were observed between isolates recovered from the MC and BC groups with respect to antibiotic resistance. AMP resistance was identified in 17·6% and 6·2% of MC and BC isolates, respectively; this was not statistically significant (P = 0·1233).
Table 3. Drug resistance level in Shigella sonnei isolates from children seeking medical care (MC) and community-based diarrhoeal surveillance study (BC) groups
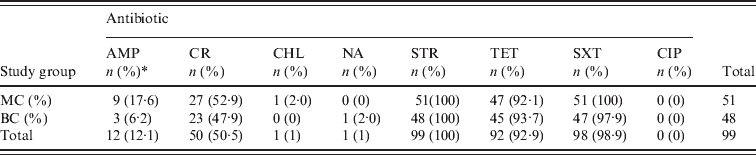
AMP, Ampicillin; CR, cephalothin; CHL, chloramphenicol; NA, nalidixic acid; STR, streptomycin; TET, tetracycline; SXT; trimethoprim–sulfamethoxazole; CIP, ciprofloxacin.
* n, Number of isolates; (%), percentage of total S. sonnei isolates resistant to the selected antibiotic.
Plasmid profile analysis
Analysis of extracted plasmid DNA revealed that all 99 S. sonnei isolates harboured multiple plasmids; 15 distinct but closely related plasmid patterns were identified (data not shown). All isolates shared a high-molecular-weight band representing the invasion plasmid and 96% of the isolates shared five other plasmid bands. No major differences were observed between the two study groups. We did observe differences between isolates recovered at different times of the study; older isolates appeared to carry unique plasmids which were not evident in more recent isolates.
Molecular analysis
All S. sonnei isolates were positive for ipaH and class II integrase genes but negative for set and class I integrase genes. Of the 99 isolates of S. sonnei, 81 (82·2%) harboured the sen gene; no significant difference in frequency was observed among isolates collected: 43 (84·3%) and 38 (79·1%) in MC and BC studies, respectively.
To determine the phylogenetical relationship of S. sonnei in isolates from the two studies, PFGE using the restriction enzyme XbaI was performed (Fig. 1). Data obtained revealed the presence of 40 closely related PFGE patterns consisting of 16–20 reproducible DNA fragments, ranging in size from about 452·7 kbp to 28·8 kbp. Three major clades (I, II, III) were apparent at a similarity value of 52%. Multiple branches were evident within each of these overarching clusters. Clade II (n = 61) was the largest and most diverse, with four distinct clusters or pulsotypes apparent at 77% similarity. Clade I (n = 31) had three clusters at this similarity value and clade III had only a single cluster. Only clade III (n = 7) was homogeneous, consisting of all isolates recovered from MC study samples. The maximum band differences separating any isolate in each cluster was four bands; consistent with the interpretation of clonal lineages as outlined previously [Reference Tenover39]. Of the eight clusters identified, three (clusters 2, 3, 6) were populated exclusively by S. sonnei isolates from one source while the remaining clusters were mixed with isolates from both studies.

Fig. 1. Shigella sonnei phylogenetic tree obtained using XbaI macrorestriction analysis Abbreviations: HS (S. sonnei isolates from Abu Homos medical clinic, Nile Delta); MH (S. sonnei isolates from Manshayet Nasser Medical Clinic in Mokattam Hills, Cairo), part of the children seeking medical care (MC) group; WS and SC, children enrolled in the Abu Homos birth cohort (BC) studies.
The majority of isolates were located in clade II (clusters 4, 5 and 7) and clade I (cluster 1). Cluster 4 contained the most isolates (n = 27) and four branches were evident at 90% similarity. One branch, containing only MC isolates from Mokattam Hills from 2005 (cluster 4, see asterisk in Fig. 1), was populated by AMP-resistant S. sonnei. The remaining branches were populated by isolates from both study populations. The largest of the branches consisted of isolates from multiple years but with an indistinguishable XbaI pattern. Clade I (cluster 2) and clade II (cluster 6) were populated with isolates exclusively from the BC study, cluster 1 was populated by 19 isolates recovered from both studies and two branches were evident with 90% similarity. One of these branches contained four similar isolates that were exclusive to the BC study. Three of these isolates were recovered during the same time period and possibly represent a mini-outbreak.
DISCUSSION
The factors affecting the emergence or decline of epidemic shigellosis are not clear. Recently, the World Health Organization emphasized the need to understand the disease burden and epidemiology of Shigella infections in developing countries [Reference Farshad40, 41]. We have been interested in the integration of epidemiological data with that of the advanced characterization of the specific Shigella spp. pathogens associated with diarrhoeal disease in Egyptian children. Previously, we described the clinical presentation, epidemiology, phenotypical and genetical characterization of S. flexneri, S. boydii and S. dysenteriae isolates acquired from children aged <5 years presenting for diarrhoea from three sites in lower Egypt [Reference Ahmed42, Reference El-Gendy43]. To date, a similar analysis of S. sonnei isolates from Egypt has not been performed. We investigated two sources of S. sonnei, isolates cultured from children seeking medical treatment – interpreted as having severe diarrhoea – and children enrolled in a birth cohort surveillance of diarrhoea. Microbiological testing demonstrated that all of the S. sonnei isolates in this study belonged to biotype g and carried a class 2 integron; this configuration is currently the globally dominant S. sonnei biotype. Clinical findings suggested that children seeking medical treatment had more severe illness than those who were identified through weekly household surveillance, although the latter group were reported to be more significantly dehydrated. However, antimicrobial susceptibility testing, plasmid profiling, virulence gene detection and PFGE provided little additional evidence to explain the increased virulence observed.
Although the S. sonnei isolates included in this study were obtained over a period of 7 years from two different sites in Egypt, biotype g was the only biotype recovered. In Italy, S. sonnei periods have been divided into an endemic phase associated with biotype a, and an epidemic phase, where clusters of biotype g emerged in more recent decades [Reference Mammina23]. These observations imply that biotype g epidemic strains have emerged and disseminated globally in recent years and highlight the current predominance of this biotype worldwide.
Our study, like previous studies in Egypt, observed S. sonnei predominance during warm months and a higher incidence in children aged >1 year [Reference Abu-Elyazeed6]. The statistically significant gender bias of S. sonnei isolated from male children seeking medical treatment is potentially explained by a family caring bias for males with respect to medical care [Reference Yount44].
Antimicrobial resistance has emerged as a major public health problem worldwide [Reference Ahmed42]. In developing countries, antibiotic resistance has been linked to the inappropriate use of antibiotics, over-the-counter availability of antibiotics, lack of healthcare personnel with adequate training, poor-quality drugs and poor sanitary conditions [Reference Putnam45]. Additionally, multi-resistance to the antimicrobial agents used in the treatment of shigellosis has been reported in many parts of the world [Reference Ashkenazi46–Reference Lee48]. The majority of our isolates were multidrug resistant (MDR) against commonly used antibiotics such as SXT, TET and STR. The frequency of resistance reported for isolates from 1999 to 2005 exceeded that observed in the preceding 5 years, and the trend for multidrug resistance was more pronounced [Reference Putnam45].
Our study showed universal carriage of class 2 integrons within S. sonnei and the complete absence of class 1 integrons, in agreement with other studies [Reference DeLappe37, Reference McIver49–Reference Pan51]. Class 2 integrons, members of the Tn7 family, harbour dfrA1, sat and aadA1 gene cassettes, conferring resistance to trimethoprim, streptothricin and spectinomycin/STR, respectively. Emergence of biotype g, class 2 integron-carrying S. sonnei can be documented to the late 1980s, and provides an explanation of the increase in resistance to trimethoprim and STR over the last two decades [Reference Mammina16]. Resistance to TET, STR and sulfa-compounds in S. sonnei has been linked to carriage of an 8·4 kbp non-transferable R plasmid carrying the genes tetA, strA-strB and sul1 [Reference Seol52]. Resistance to AMP was higher in the MC group than in the BC group (17·6% vs. 6%); however, it should be noted that 7/9 isolates showing resistance to AMP in the MC group were obtained from the same site, within the same time period, and showed the same genetic lineage (cluster 4). These data suggest that an outbreak of S. sonnei might have occurred in this location. Future diarrhoea-based studies in Egypt should incorporate as close to real-time PFGE sample analysis and epidemiological follow-up to determine whether identified clusters are actual outbreaks, consistent with the goals of PulseNet [Reference Gerner-Smidt53]. Only a few isolates were resistant to CHL and no resistance was observed to CIP; we speculate that this was probably because of the infrequent use of these drugs as alternative therapeutic regimens in Egypt. Our study does document, for the first time, NA-non-susceptible (one resistant and four intermediate isolates) S. sonnei in Egypt. Interestingly, our NA non-sensitive isolates were from different field sites, which might imply an emergence of NA-resistant strains or possible spread between population groups.
In the present study, plasmid profiling indicated that S. sonnei isolates shared a high level of similarity; 15 closely related plasmid patterns were identified. However, plasmid profile analysis was insufficient to distinguish differences between S. sonnei isolated from children seeking medical care and those participating in the BC study. Differences between the plasmid profiles of isolates recovered at different times of the study were more obvious, potentially serving as a useful marker for strain evolution over time [Reference Farshad40]. While it remains possible that plasmids may have been lost from bacterial isolates after long-term storage [Reference Navia, Gascon and Vila54], we note that isolates maintained the same antibiotic resistance pattern over the time of the study, suggesting plasmid loss was unlikely.
Shigella spp. produce enterotoxin 1 (ShET-1) and enterotoxin 2 (ShET-2) [Reference Nataro9, Reference Noriega55]. We found no evidence of the set gene in our S. sonnei collection, in accord with other reports [Reference Talukder5, Reference Nataro9, Reference Farfán56]. The sen gene (encoding ShET-2) has been reported to range from 10% to 58% prevalence in isolates of S. sonnei [Reference Talukder5, Reference Vargas33, Reference Roy57], although it has been shown [Reference Vargas33] that all isolates recovered during specific time periods carry the toxin gene. In this study, 82·2% of S. sonnei harboured the sen gene (84% and 79% in MC and BC samples, respectively). It is possible that the virulence plasmid was lost in the 18 S. sonnei isolates in which set was not detected, due to laboratory passage and the age of the cultures. It appears, however, that S. sonnei isolated from children in Egypt are more often associated with the sen gene than S. sonnei from other studies.
PFGE is a useful tool for routine subtyping of bacterial isolates, leading to the detection of clusters of infection as well as changes in population structure [Reference Ribot36, Reference DeLappe37, Reference Chu58]. Although PFGE analysis revealed the presence of closely related S. sonnei isolates, there was limited discrimination between isolates from the MC and BC studies. Several S. sonnei pulsotypes were observed circulating through our study populations over the time of the study. Although some pulsotypes were specific to a certain time period, others appeared to persist throughout the study. For instance, several branches in clade II (cluster 4) consisted of isolates from different years; whereas two of the branches in clade I (cluster 1) only had isolates from the same time period. Interestingly, the largest branch in clade II (cluster 4), consisting of indistinguishable isolates, persisted over time. In Egypt, several S. sonnei biotype g clones were dominant over the time of the study; however, only a few pulsotypes persisted over time. A comparable observation has been reported in Italy and Korea [Reference Mammina23, Reference Jin24]. One of the limitations of this study is the application of a single phylogenetic method to characterize the isolates. Advances in alternative genetic techniques such as MLVA [Reference Liang28, Reference Chiou29] might provide increased resolution to studying S. sonnei populations in future studies.
In conclusion, our study was intended to provide baseline data regarding the population structure of S. sonnei in Egypt. We selected isolates recovered from children aged <5 years from two separate studies representing severe diarrhoeal infection (MC) or for mild to moderate diarrhoea (BC). Clinical symptoms supported the increased severity in disease associated with MC cases, but the laboratory data in most instances did not clearly distinguish between sample sources. The explanation for this could be variations in host immune response between the patients or to the low discriminative power of PFGE. Future studies will need to utilize more advanced molecular tools such as MLVA to evaluate population homogeneity. Increases in MDR cases and the observation of NA-resistant isolates of S. sonnei argue for establishment of a national surveillance programme to monitor changes in S. sonnei antibiotic resistance.
ACKNOWLEDGEMENTS
This study [DoD no. NAMRU3.2000.0002 (IRB Protocol no. 2000.0002.96), IRB no. NAMRU3. 1997.0002.72, IRB no. NAMRU3.1998.0003.75, IRB no. 145 (DOD no. NAMRU3.2003.0011)] was reviewed and approved by the Institutional Review Boards of the U.S. Naval Medical Research Unit No. 3, and the Egyptian Ministry of Health in compliance with all Federal regulations governing the protection of human subjects. Informed consents were obtained from all adult participants and from parents or legal guardians of minors.
DECLARATION OF INTEREST
None.








