INTRODUCTION
New, emerging or re-emerging infections pose a constant threat to society and systems to assess the level of threat are essential. An average of more than three new human pathogen species have been discovered each year since 1980, the majority of which are zoonoses [Reference Woolhouse and Gaunt1, Reference Woolhouse and Gowtage-Sequeria2]. A range of activities have been developed to attempt to identify new infectious threats as they emerge in order to facilitate a timely response including control [Reference Kaiser3–Reference Paquet5].
In England, the National Expert Panel on New and Emerging Infections (NEPNEI) was established following the publication of ‘Getting ahead of the curve: a strategy for infectious diseases’ [6] by the Chief Medical Officer in 2002. The remit of the Panel includes identifying ‘key areas for action and to advise on priorities by … identifying emerging and potential infectious threats to public health both nationally and internationally’ [7]. Our group undertakes horizon scanning activities to identify such potential infectious threats on behalf of the Panel. A systematic process is used to gather information about current outbreaks or incidents of potential new and emerging infectious diseases in animals or humans, occurring anywhere in the world. A wide range of sources are reviewed daily, including news and surveillance reports, scientific literature, scientific search engines, and journals. Potential threats are reported to the Panel and others working in health protection, and as this process developed it became clear that a rapid assessment of the risk posed by such threats was necessary to inform the level of response required.
Risk assessment is an integral part of the wider process of risk analysis, which starts with hazard identification, and includes risk management and communication. A considerable volume of literature exists on risk assessment, particularly focusing on the nuclear, aviation, chemical, automotive and food industries. A common approach is the amalgamation of a range of objective and subjective data to produce risk-related standards. Events which are on-going, unfolding or ‘one off’ must be considered in the assessment of a specific threat. Many of these techniques can be adapted and applied to public health, where it is particularly important to recognize the potentially significant consequences of seemingly small risks.
The purpose of this work was to develop a rapid, systematic, objective and transparent method for assessing the risk to the UK population from new and emerging infections arising anywhere in the world. In this context, risk is a product of the chance that a new or emerging infection (either human or zoonotic) will infect the UK human population, and the impact this will have on human health. This process is important to identify and inform risk management action and intervention, for example making recommendations for enhanced surveillance, implementation of control measures or urgent establishment of an expert group to deal with a specific problem. The work also provides a means of communicating risk in a systematic manner to stakeholders and provides an audit trail for decisions made.
METHODS
Previous works on risk assessment relevant to public health were reviewed to define best practice and consistency of approach. Although the literature on risk assessment is extensive, there is little that is specific to emerging public health threats. Two major projects were identified.
The first, ‘HPZone’, was a Department of Health (England) funded project to develop a risk management model for managing communicable disease incidents [8]. This project drew on expertise in risk assessment from industry to develop a decision support tool based on five key aspects of risk. These include severity, confidence in diagnosis, spread, interventions (available), and (political and public) context. Each aspect is scored on a scale of 0–4 and the scores combined to produce an overall risk score. This tool has been developed and tested in a number of workshops around the country, and was found to enhance clinical decision-making.
The second project was devised by the Department for the Environment and Rural Affairs (Defra) as part of the prioritization goal of their Veterinary Surveillance Strategy [9]. A risk assessment model has been developed in which probability and impact are considered separately, and which uses weighted criteria to quantify risk and provide justification as to why a disease or animal population is/is not considered to warrant government intervention [10]. Threats/diseases can then be ranked in order of resource priority. Impact is considered separately in relation to public health, animal welfare, international trade and the effects on wider society and the economy.
Based on the findings from these two projects, a risk assessment tool was developed which utilizes algorithms comprising a series of decision questions in order to derive a qualitative estimation of risk. The algorithms which formed the basis for the risk assessment tool were piloted for 6 months and then endorsed by NEPNEI. They have been used to assess the risk to the UK from a range of infections identified by our horizon scanning activities. The initial risk assessment process is undertaken by a scientist using a variety of sources of information, including published scientific literature, formal and informal reports and expert opinion. The document is then passed to the Human Animal Infections and Risk Surveillance group, a multidisciplinary group of human and animal health experts [Reference Walsh and Morgan11] for comment, discussion and approval before going to the NEPNEI for review and sign-off. The nature of emerging infections means that information is often lacking, and the risk assessments must be revised in light of new developments, or if the information on which they are based changes in any way.
RESULTS
Separate algorithms were developed to consider the probability and impact of the emerging infectious threat, because combination into a single parameter of ‘risk’ would have resulted in loss of crucial information.
Probability of infection
The possibility of an infectious threat causing infection in the UK human population
This depends on a number of factors including the introduction and establishment of a known human or zoonotic disease into the UK, the presence of appropriate environmental conditions to support the natural reservoirs/vectors of disease, and contact with susceptible individuals (Fig. 1). If the zoonotic potential is unknown, then this is first assessed using a process reported previously [Reference Palmer, Brown and Morgan12]. As this is a qualitative process a numerical probability is not assigned, probability in this context is used in a general sense.
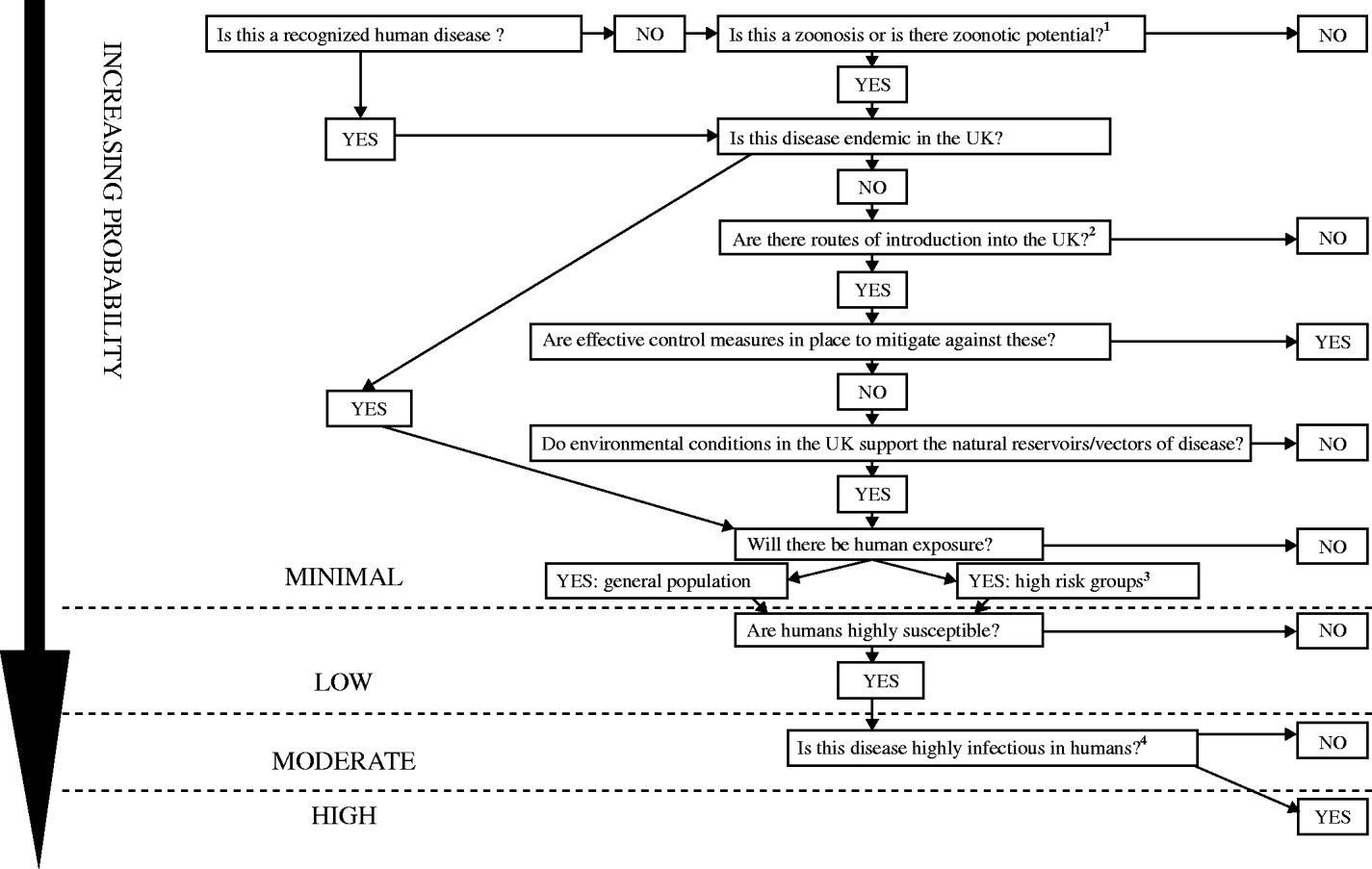
Fig. 1. Probability of infection: the likelihood of an infectious threat causing infection in the UK human population. 1Zoonotic potential: (refer to zoonotic algorithm [7]) the potential for an infection to be naturally transmissible from vertebrate animal hosts to humans. The causative organism may be viral, bacterial, fungal, protozoan, or parasitic. Transmission may be direct (in occupational settings or due to leisure activity or to keeping pets) or via food and water. The following questions should be considered:
• Is the infection pathogenic in humans?
• Is there evidence of cross species transfer?
• Is there serological evidence of infection in humans?
• Is there documented human disease or an equivalent disease in humans?
• Food
• Animal
• Bird
• Human
• Travellers (check geographical distribution – areas where UK travellers visit)
• Occupational groups (degree of contact between host species and humans)
• Specific age groups (e.g. children/elderly)
• Other groups – specify (infection specific, e.g. recreational/healthcare workers)
• Mode of spread
• Period of communicability
• Length of incubation period
• Length of asymptomatic infection
Impact on human health
The scale of harm caused by the infectious threat in terms of morbidity and mortality
This depends on mode of transmission (whether person-to-person, zoonotic or vector-borne), infectiousness, severity of illness caused and availability of interventions (Fig. 2).
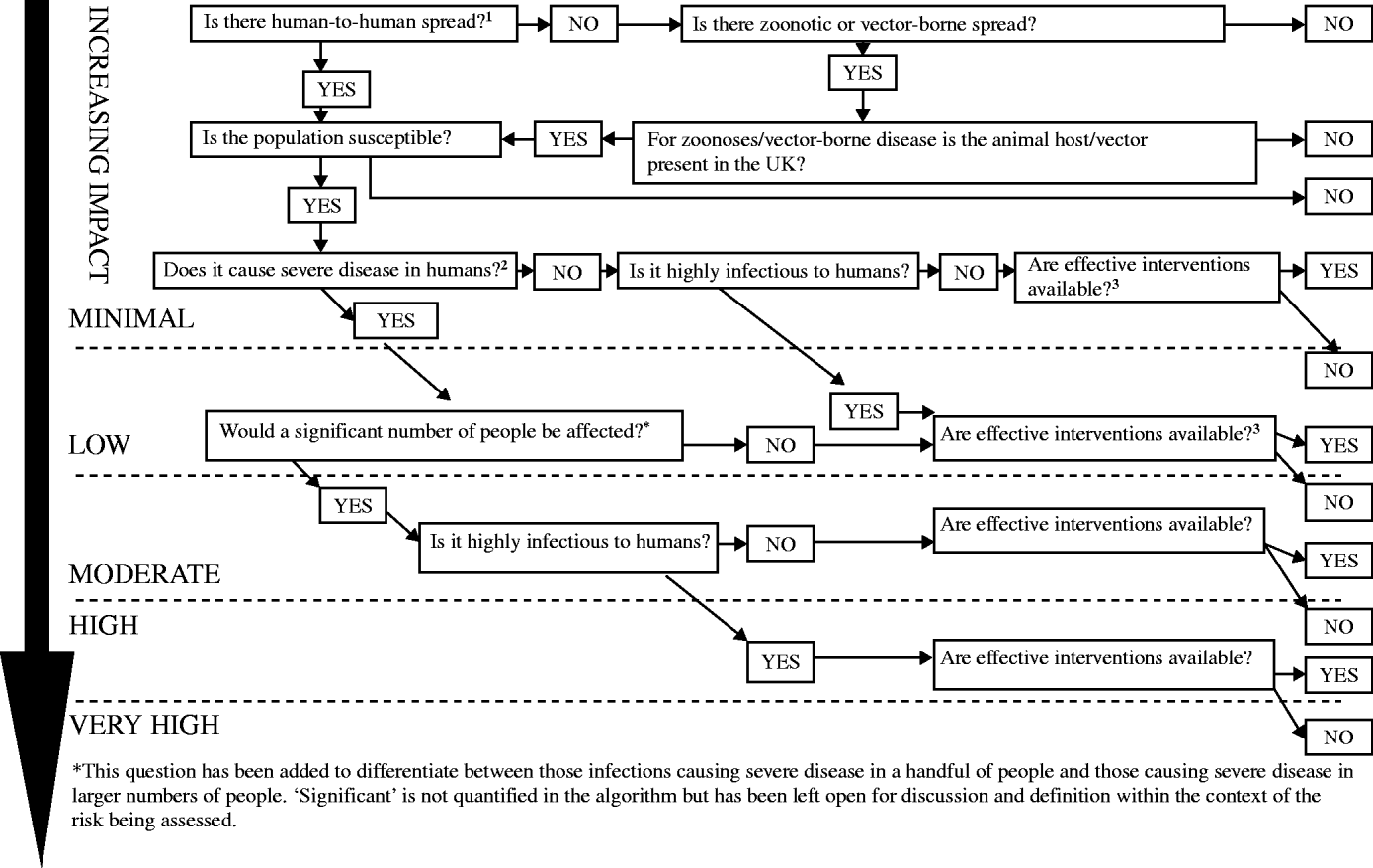
Fig. 2. Impact on human health: the scale of harm caused by the infectious threat in terms of morbidity and mortality. 1Spread is the intrinsic temporal and spatial potential for spread including the infective dose, the virulence of the organism, the availability of the route of spread, the observed spread and the susceptibility of the population in the UK. 2Severity: the seriousness of the disease in terms of the intrinsic propensity in the specific circumstances to cause harm to individuals or to the population. Includes:
• Morbidity, mortality and case fatality
• Complications
• Burden of disease in terms of contacts with health services (e.g. calls to NHS Direct, GP consultations, A&E attendances, hospital admissions).
• Is effective treatment available?
• Is prophylaxis available?
• Are logistics in place to deliver these?
The algorithms make use of all available information (scientific, clinical, epidemiological, and veterinary) in assessing the level of risk, and they also aid in the identification of gaps in knowledge. For a new agent, such gaps may be significant and these are clearly documented in the assessment. It is difficult to rapidly assess potential threats where much of the information necessary to inform the risk process is not known, and this uncertainty is managed in the algorithms by always adopting a precautionary approach and moving through the algorithm to a higher level of risk. Any uncertainty can be reflected as a hatched area in the final algorithm (see worked example below).
Context
Another factor which may affect the perception of risk is the context in which a potential threat arises (see Fig. 3). Here context is taken to mean the societal environment in which events are occurring and decisions on responses are being made. The assessment incorporates public concern and expectations, professional knowledge and external factors such as politics and economic impact. Context is considered separately as it is difficult to assess; however, it is important to consider, as infections assessed as low risk could have significant economic and political outcomes. The requirement for a public health response and any issues likely to affect the risk process are highlighted on the front page of the risk assessment.

Fig. 3. Effect of context on risk. Context: the broad environment, including public concern and expectations, professional knowledge and external factors including politics, in which events are occurring and decisions on responses are being made. Many of the factors leading to interpretation and communication of risk are complex and often difficult to predict. However, these may over-ride all scientific evidence and require a response which may be disproportionate to the actual risk. The following factors may influence assessment of risk beyond probability and impact. Factors that may distort or attenuate the perception of risk:
• Lack of professional knowledge about disease epidemiology
• Conflicting professional opinion
• Significant uncertainty including outcome
• Severity, numbers affected, case fatality
• Perceived vulnerability of those affected (e.g. pregnant women and children)
• Lack of available treatment/interventions
• Significant public concern
• Significant political interest
• Significant media interest
This risk assessment tool is intended to be qualitative rather than quantitative and therefore an overall numerical score is not assigned to each infection. Instead infections are assigned a level of risk (minimal, low, moderate, or high) for probability, and impact (see Fig. 4), in order to inform further action and urgency of response. Resulting risk assessments are retained for reference and revised as the situation changes or new evidence arises. The assessments are based on current knowledge and do not attempt to predict future trends; however, the risk assessments are updated and revised as the situation changes or new information arises. Each new version is numbered and dated accordingly, with the new information clearly marked.

Fig. 4. Levels of risk.
USING THE RISK ASSESSMENT TOOL: WORKED EXAMPLE
Chikungunya infection
Chikungunya is a vector-borne viral infection which was first identified as a potential hazard in March 2005, when outbreaks were reported on a number of islands in the Indian Ocean, popular destinations for UK travellers. The situation was monitored until March 2006 when it was considered to be a potential risk to the UK and a risk assessment was carried out. The risk assessment was subsequently updated in May, June and November 2006, twice during 2007, and again during 2008, as further information emerged and the situation could be re-evaluated.
Probability
Please read in conjunction with the Probability Algorithm (Fig. 5) following the boxes shaded grey. Where the evidence may be insufficient to give a definitive answer to a question the alternative is also considered with the most likely outcome shown in solid grey and the alternative outcome in grey hatching.
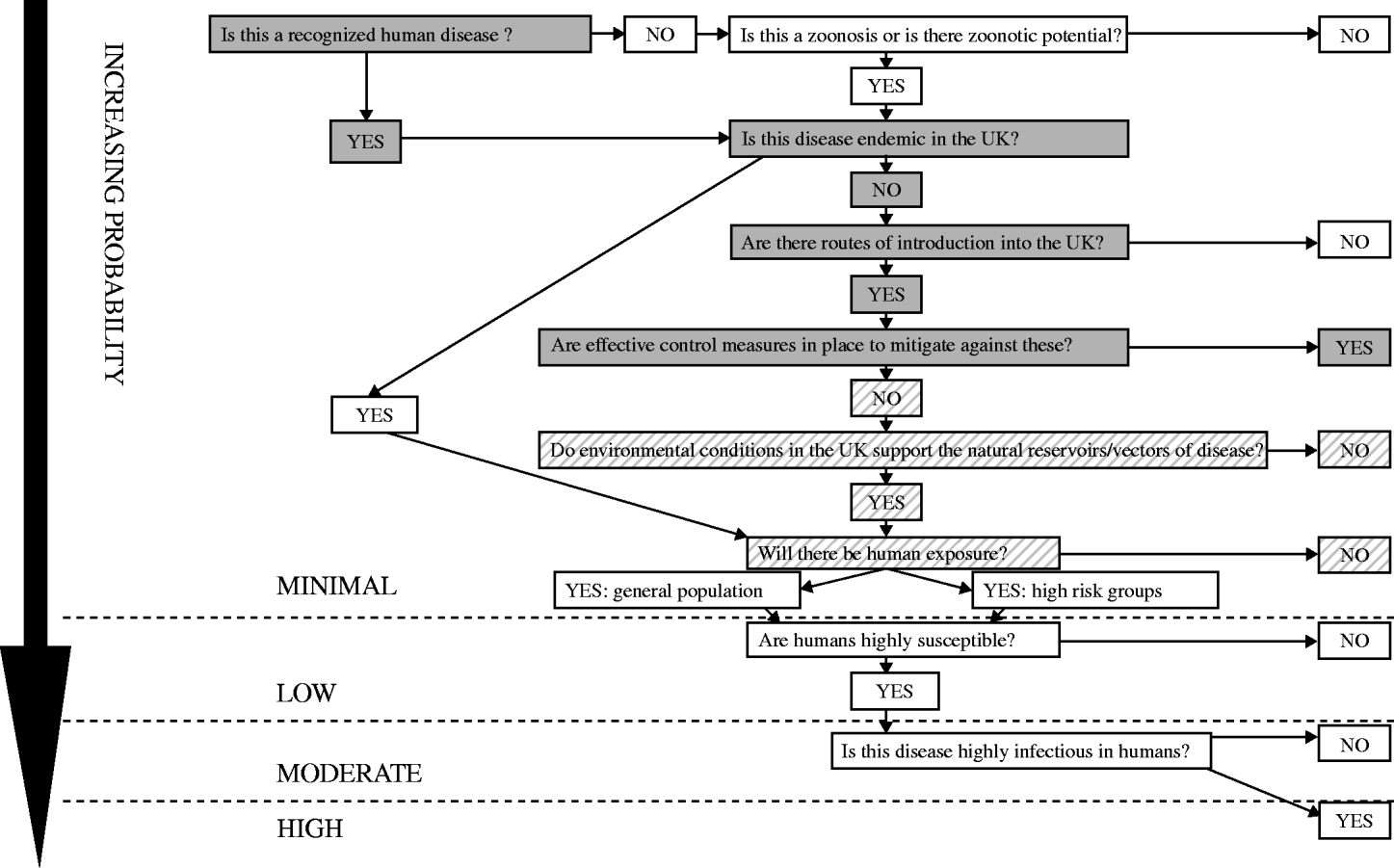
Fig. 5. The probability of human infection with chikungunya virus in the UK population: minimal.
Is this a recognized human disease? Yes. Chikungunya virus is an arbovirus first isolated in Tanzania and Uganda in 1953. The main reservoirs are monkeys, but other species can also be affected including humans. The virus is transmitted from human to human by Aedes mosquitoes (A. aegypti and A. albopictus in Asia and the Indian Ocean, with transmission by a wide range of Aedes species in Africa) [Reference Pialoux13].
In 2005, an outbreak of chikungunya fever began in the Comoros Islands, was then reported in Mauritius and Mayotte, and finally in the island of Réunion, an overseas administrative département of France, in March 2005. Between 1 March 2005 and 30 April 2006 about 255 000 cases of chikungunya infection were reported on Réunion. The majority of cases occurred after December 2005, with a peak of 45 000 reported in the week of 29 January to 4 February 2006.
Although chikungunya is generally not considered fatal, a study of the Réunion outbreak found that the crude death rate exceeded expected levels. An estimated 260 excess deaths on the island were attributed to chikungunya infection from January to April 2006, the majority in patients aged ⩾75 years [Reference Josseran14].
Probable mother-to-child transmission of the virus was also reported for the first time during this outbreak [Reference Ramful15].
Between February and October 2006 about 1·36 million cases of chikungunya were reported across 151 districts of India; in the provinces of Andhra Pradesh, Andaman & Nicobar Islands, Tamil Nadu, Karnataka, Maharashtra, Gujarat, Madhya Pradesh, Kerala and Delhi. In some areas reported attack rates reached 45% [Reference Pialoux13].
Is this disease endemic in the UK? No.
Are there routes of introduction into the UK? Yes. Many of the areas where chikungunya virus is endemic are popular tourist destinations and the virus may be imported into the UK in a viraemic traveller. Onward transmission would require the presence of competent vector mosquitoes. The initial viraemic phase of the disease lasts for a week after onset of symptoms [Reference Pialoux13, Reference Ramful15], and transmission could occur through blood transfusion from an infected donor.
Between March 2005 and February 2006, 140 cases of chikungunya were imported from Réunion to France. As the vector A. albopictus has also been identified in limited areas in southern France there is some concern over the risk of onward transmission from imported cases. One autochthonous case was reported in metropolitan France, in a nurse following a probable blood exposure incident while caring for a patient with an imported infection [Reference Cordel16, Reference Parola17]. Cases of chikungunya were also imported to other European countries including Germany, Switzerland, Italy and Norway from the affected islands in the Indian Ocean [Reference Pfeffer and Loscher18].
The Health Protection Agency's Special Pathogens Reference Unit reported 133 cases with evidence of chikungunya infection in the UK in 2006. These include 45 laboratory-confirmed cases (PCR and/or virus isolation), 30 probable cases (clinical symptoms and IgG, IgM positive), 35 suspected cases (clinical symptoms, relevant travel history and IgG positive), and 23 with past exposure (IgG positive with no clinical details or travel history, or IgG positive with history of past exposure). Of these cases, 68 reported travel to the Indian Ocean islands and 34 to India. Cases were also reported in UK travellers returning from Africa and Australia [19].
An outbreak of chikungunya fever was confirmed in the province of Ravenna, northeast Italy in 2007, with a total of 205 cases confirmed between 4 July and September 27 in the villages of Castiglione di Cervia and Castiglione di Ravenna. The presumed index case was a traveller from India, who developed symptoms while visiting relatives in the village where the outbreak began. A. albopictus is present in Italy and this outbreak provides the first evidence of transmission of chikungunya virus by local mosquitoes in Europe [Reference Rezza20]. Autochthonous transmission is also thought to have occurred in the city of Bologna, 75 km from the original cluster. Two cases were reported in September 2007 in women resident in Bologna, with a history of recent travel to the affected areas in Ravenna. A further three cases were reported in December 2007 in Bologna residents living on the same floor of a building 2·5 km from the previously identified cases, with no reported history of travel to the affected areas at the time of the outbreak [Reference Seyler21].
Are there effective control measures in place to mitigate against these? Yes Footnote †. Travel advice on mosquito bite avoidance has been issued to those visiting affected areas in the islands of the Indian Ocean and mainland India. Control of chikungunya in endemic areas is based on vector control, particularly the reduction of mosquito breeding sites (removal of containers with stagnant water or treatment with larvicides) and elimination of adult mosquitoes using insecticides.
Do environmental conditions in the UK support the natural reservoirs/vectors of disease? Yes†. In Asia and the Indian Ocean region the main chikungunya virus vectors are A. aegypti and A. albopictus [Reference Pialoux13]. Transmission in Africa involves a sylvatic cycle, primarily with A. furcifer and A. africanus [Reference Powers22].
Thirty-three species of mosquitoes have been recorded in the British Isles, including A. cinereus, A. vexans, at least 10 Ochlerotatus species and one Finlaya species – (all formerly Aedes); however, none of the Aedes species implicated in the transmission of chikungunya virus have been found. Only A. albopictus, which has been found in other European countries but not so far in the UK, has the theoretical potential to survive in the UK climate. Chikungunya virus is unlikely to become established in the UK in the absence of the main vectors [Reference Medlock, Snow and Leach23, Reference Medlock24].
Will there be human exposure? No. Currently there would be no human exposure in the UK as A. albopictus is not known to be established. There have been no authochthonous cases of chikungunya infection in the UK.
The PROBABILITY of human infection with chikungunya virus in the UK population: MINIMAL.
Impact
Please read in conjunction with the Impact Algorithm (Fig. 6) following the boxes shaded grey.
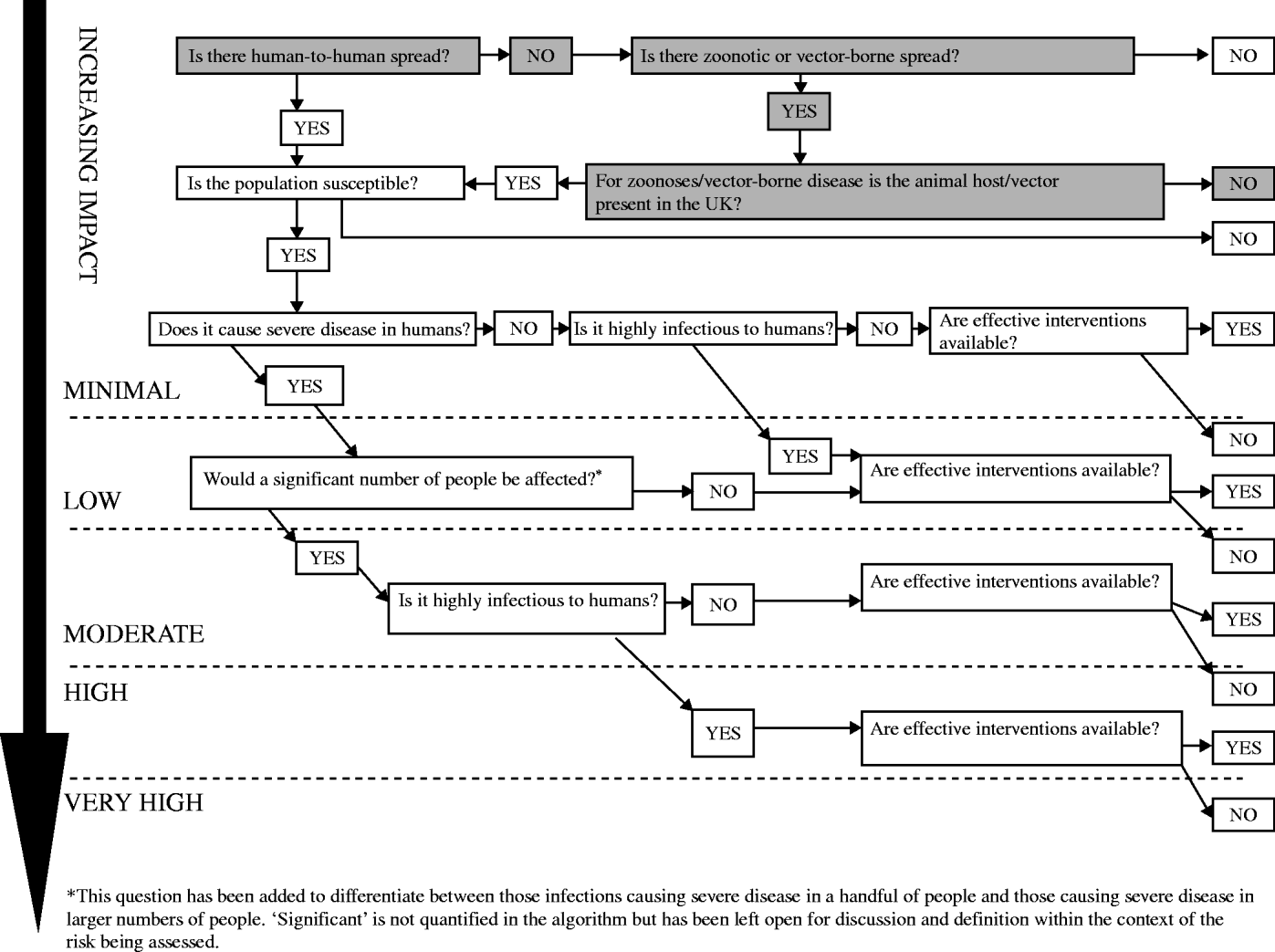
Fig. 6. The impact of chikungunya virus on human health in the UK: minimal.
Is there human-to-human spread? No. Chikungunya virus is a vector-borne virus. Human-to-human transmission has not been reported; however, there was some evidence of mother-to-child transmission in the outbreak on Réunion Island. A diagnosis of chikungunya infection was confirmed in six newborns that showed symptoms of acute infection and presented with a clinical picture of meningoencephalitis within 5 days of birth. The mothers of all six children had acute chikungunya infection within the 48 h before delivery. The incubation of chikungunya virus is 2–4 days (range 2–10 days) [25, Reference Paquet26].
Is there zoonotic or vector-borne spread? Yes. Chikungunya virus is an arbovirus (arthropod-borne virus) and the primary vector is A. aegypti; however, A. albopictus was implicated as the main vector in the large outbreak on Réunion. High viral loads in patients returning from Indian Ocean islands to countries where A. albopictus is prevalent could lead to onward transmission and potential outbreaks [Reference Parola17].
For zoonoses/vector-borne disease, is the animal host/vector present in the UK? No. Although A. albopictus has been found in other European countries none of the Aedes species implicated in transmission of chikungunya virus (including A. albopictus) are currently found in the UK [Reference Medlock, Snow and Leach23, Reference Medlock24].
The IMPACT of chikungunya virus on human health in the UK: MINIMAL.
Context
There is no current reason to believe that the context might significantly affect the perception of risk of chikungunya virus in the UK. However, the situation will be monitored and the risk assessment reviewed as more information becomes available.
DISCUSSION
We have devised and piloted a simple risk assessment tool to rapidly assess the potential risk of new, emerging or re-emerging infections to the UK population. This has been used to assess the threat of a range of infections, and communicate this information to scientists, policy makers, government departments and other agencies.
The tool provides a standardized framework for estimating risk, and identifies not only current gaps in knowledge but also the need for an expert formal risk assessment. In addition it acts as a log of decisions made and actions taken at specific time points. An advantage of this approach is that it generates an audit trail, which records how assessments have been reached and the information that was available at the time. The evidence used at each stage of the decision tree is fully referenced, allowing the reader to study an issue further where necessary. The outcome of this process can be used to promote risk-informed decision-making, and ensure that complex decisions become easier to explain and justify at each stage of the decision process.
A formal risk assessment of this nature should be considered for any new or emerging infection in humans or animals, unless there is good evidence that the infection is neither a recognized human disease nor a potential zoonosis. Where a new agent is identified there may be insufficient information to carry out a risk assessment and this should be clearly documented. The risk assessment tool is not intended to replace the definitive, expert-based, weighty process that can be carried out with more information, but rather as a rapid ‘first-sift’ process, highlighting gaps in knowledge and indicating whether a new threat warrants urgent action or not. This tool is qualitative rather than quantitative and potential infectious threats are assigned a level of risk (minimal, low, moderate, high) for probability and impact. There are several reasons for this approach. First, it may not be possible or accurate to give a quantitative score to infections for which there is little available information. Second, this approach allows differentiation between high-impact, low-probability events and those which are more common but less severe. This information is crucial for risk management and would be lost by combining scores into an overall risk score.
This system allows risk to be assessed rapidly and early in an incident. This is particularly appropriate for emerging infections, as a lack of information is a common feature of many incidents. Qualitative risk assessment allows a threat-specific interpretation to be made, and the inclusion of context as a parameter adds further unquantifiable information to the assessment. Context is often difficult to predict and relevant special features of the incident are noted on a summary front sheet. The collation of information on a new threat into one document aids risk communication and ensures that those responsible for risk management are aware of the evidence behind the advice given.
The risk assessments are initially undertaken by members of the Human Animal Infections and Risk Surveillance group [Reference Walsh and Morgan11]. Since the majority of emerging infections are zoonoses, we feel a multidisciplinary approach is essential and collaboration between experienced colleagues working in human and animal health, disease surveillance, research and policy making allows a thorough consideration of the different aspects of a specific hazard. This work has become an important component of our threat and risk assessment activities. Horizon scanning identifies potential threats and the application of the rapid risk assessment allows us to alert and communicate with policy makers and those involved in health protection in an explicit, standardized way.
The process is simple enough to be relatively transparent, acknowledges uncertainty where it exists and is flexible enough to apply to a very wide range of infectious threats. This risk assessment tool has been applied to a range of infections including Chikungunya virus, hantavirus, Japanese encephalitis and avian influenza, and has proved invaluable in documenting and communicating risk to a range of stakeholders.
A number of threat reporting and alerting systems [Reference Paquet5, Reference Brownstein27–Reference Grein30] have been established in recent years, which generate a great deal of information. This tool allows threats of varying severity to be rapidly assessed and filtered, ensuring that those requiring immediate action can be dealt with.
ACKNOWLEDGEMENTS
The authors thank the following people for their contributions: Professor Angela MacLean for reviewing the manuscript; the members of the Human Animal Infections and Risk Surveillance Group for their support and contributions to the risk assessments, including Paul Gayford, Robert Smith, David Brown, Jolyon Medlock and Geoff Pritchard for their input into the development of the process.
DECLARATION OF INTEREST
None.








