Introduction
Protoporphyrinogen IX (PPO)–inhibiting herbicides have been used for weed control in many row crops for over 50 yr. Many troublesome broadleaf weeds, particularly weeds resistant to acetolactate synthase inhibitors and glyphosate, are controlled by PPO inhibitors applied PRE and POST in soybean [Glycine max (L.) Merr.] and cotton (Gossypium hirsutum L.). In recent years, PPO resistance (PPO-R) in Palmer amaranth has been in confirmed in Arkansas, Tennessee, and Illinois in 2011, 2015, and 2016, respectively (Heap Reference Heap2018).
Waterhemp [Amaranthus tuberculatus (Moq.) J. D. Sauer] (syn. rudis) was the first weed species reported to be resistant to PPO-inhibiting herbicides (Heap Reference Heap2018). To date, PPO-R waterhemp has been well documented and infests most of the midwestern United States (Heap Reference Heap2018). The most common mechanism of resistance in PPO-R waterhemp is a codon deletion of a glycine residue at position 210 (ΔG210) of a PPO gene (Patzoldt et al. Reference Patzoldt, Hager, McCormick and Tranel2006). This deletion destabilizes the α-8 helix-capping region, unraveling the last turn of the helix, which enlarges the active-site cavity by about 50% (Dayan et al. Reference Dayan, Daga, Duke, Lee, Tranel and Doerksen2010). Salas et al. (Reference Salas, Burgos, Tranel, Singh, Glasgow, Scott and Nichols2016) documented this same mechanism of resistance to PPO inhibitors in Palmer amaranth in Arkansas. In a statewide survey of Arkansas, researchers found that only 55% of PPO-R Palmer amaranth plants carried the ΔG210 mutation (Salas-Perez et al. Reference Salas-Perez, Burgos, Rangani, Singh, Refatti, Pivetam, Tranel, Mauromoustakos and Scott2017). Additionally, a survey of west Tennessee in 2016 (15 counties) found that only 40% of fields infested with PPO-R Palmer amaranth could be accounted for by the ΔG210 mutation (unpublished data). The ΔG210 mutation in the 2016 west Tennessee survey was detected using methods described in Wuerffel et al. (Reference Wuerffel, Young, Lee, Tranel, Lightfoot and Young2015). Subsequent to the aforementioned surveys in Arkansas and Tennessee, Giacomini et al. (Reference Giacomini, Umphres, Nie, Mueller, Steckel, Young, Scott and Tranel2017) reported two new mutations associated with PPO-R in Palmer amaranth.
In addition to the ΔG210 mutation, two new mutations that encode for a glycine (R128G) or a methionine (R128M) instead of an arginine at the 128th amino acid residue (R128) (referred to as R98 in Giacomini et al. Reference Giacomini, Umphres, Nie, Mueller, Steckel, Young, Scott and Tranel2017) have been discovered (Giacomini et al. Reference Giacomini, Umphres, Nie, Mueller, Steckel, Young, Scott and Tranel2017; Varanasi et al. Reference Varanasi, Brabham, Norsworthy, Nie, Young, Houston, Barber and Scott2017). The R128 amino acid residue is homologous to common ragweed’s (Ambrosia artemisiifolia L.) R98, where a leucine substitution conferred resistance to fomesafen (Rousonelos et al. Reference Rousonelos, Lee, Moreira, VanGessel and Tranel2012; Salas-Perez et al. Reference Salas-Perez, Burgos, Rangani, Singh, Refatti, Pivetam, Tranel, Mauromoustakos and Scott2017). The ΔG210 mutation, R128G, and R128M mutations in Palmer amaranth were identified in accessions from Arkansas and Tennessee (Giacomini et al. Reference Giacomini, Umphres, Nie, Mueller, Steckel, Young, Scott and Tranel2017). Likewise, Giacomini et al. (Reference Giacomini, Umphres, Nie, Mueller, Steckel, Young, Scott and Tranel2017) found that an accession from Arkansas exhibited segregation for both the ΔG210 and R128G mutations in different plants. After further investigation, this population from Woodruff County, AR, was shown to exhibit cross-resistance to PPO-inhibiting herbicides from five different chemical families (Schwartz-Lazaro et al. Reference Schwartz-Lazaro, Norsworthy, Scott and Barber2017).
Since the discovery of the R128G and R128M mutations, researchers have indicated the importance of identifying the specific mutation(s) within a population where cross-resistance of PPO-inhibiting herbicides is possible (Schwartz-Lazaro et al. Reference Schwartz-Lazaro, Norsworthy, Scott and Barber2017). Growers should be aware of the mutations associated within their PPO-R populations and the potential for reduced herbicide activity present within these populations. In 2017, a survey of 18 fields in west Tennessee was conducted to determine the distribution of the three PPX2 mutations associated with PPO-R Palmer amaranth. Understanding the distribution and prevalence of these PPX2 mutations could persuade growers to utilize integrated weed management strategies to avoid further herbicide resistance spread and development.
Materials and Methods
Plant Material
Palmer amaranth infestations in grower fields, ranging from 50 to 150 plants per location, were randomly selected across west Tennessee for this survey. Plants of 8 to 10 cm height were treated with 265 g ai ha–1 of fomesafen (Flexstar® 1.88 EC; Syngenta Crop Protection Inc., Greensboro, NC) plus 0.5% vol/vol nonionic surfactant (Activator 90; Loveland Products Inc., Greeley, CO) to select for fomesafen-resistant plants. Field locations, based on the geographic location within west Tennessee, were categorized as North, Central, or South region (Table 1). At 3 to 5 d after treatment (DAT), plants were scored resistant or susceptible based on response of Palmer amaranth (Table 1; Figure 1). A population was considered resistant if plants with a surviving apical meristem were present following the fomesafen application. Tissue from new leaf growth (1.5 cm2) from up to 10 randomly selected Palmer amaranth plants at each surviving population were placed into separate 1.5-ml microfuge tubes and stored at –80 C until use. Using a CTAB (cetyltrimethylammonium bromide) protocol, genomic DNA from plant tissue of surviving plants was extracted for further analysis to detect the three known PPX2 mutations (Doyle and Doyle Reference Doyle and Doyle1987). For each location, the frequency of each mutation was expressed as a percentage of the individuals sequenced within that given field. If none of the three mutations was detected within a field, the frequency was expressed as percent (%) not characterized. All maps in this paper were created using ArcMap 10.5 (ESRI, Redlands, CA).
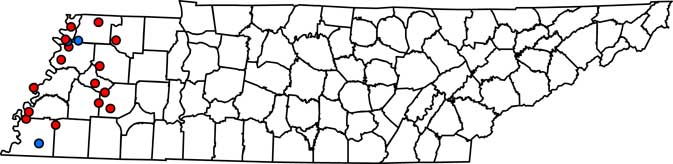
Figure 1 Field locations in west Tennessee where Palmer amaranth populations were treated with fomesafen at 265 g ai ha–1. At 3 to 5 d after treatment, fields were determined as a resistant or susceptible population. If the population was resistant, plant material from 10 plants was collected for gDNA extraction. PPO-R, PPO-resistant Palmer amaranth; PPO-S, PPO-susceptible Palmer amaranth. Red circles, PPO-R; blue circles, PPO-S.
Table 1 Location, GPS coordinates, region in west Tennessee, and response of each field screened for PPO-R Palmer amaranth.

a Abbreviations: R, PPO-resistant (field had surviving Palmer amaranth 3 to 5 d after application of fomesafen at 265 g ai ha–1); S, PPO-susceptible (100% control of Palmer amaranth 3 to 5 d after application of fomesafen at 265 g ai ha–1).
PPX2 ΔG210 Assay
The presence of the ΔG210 mutation was detected using a modified version of the Wuerffel et al. (Reference Wuerffel, Young, Lee, Tranel, Lightfoot and Young2015) TaqMan qPCR assay. The assay determines whether a plant is wild type or heterozygous/homozygous for the ΔG210 mutation using allele-specific probes (Giacomini et al. Reference Giacomini, Umphres, Nie, Mueller, Steckel, Young, Scott and Tranel2017). This modified version of the assay uses new primers that recognize both Palmer amaranth and waterhemp PPX2 sequence, PA-tqF1 (5′-TGATTATGTTATTGAC CCTTTTGTTGCG-3′) and PA-tqR1 (5′-GAGGGAGTATAAT TTATTTACAACCTCCAGAA-3′) (Giacomini et al. Reference Giacomini, Umphres, Nie, Mueller, Steckel, Young, Scott and Tranel2017).
dCAPs Assay for Detection of the R128G and R128M Mutations
Giacomini et al. (Reference Giacomini, Umphres, Nie, Mueller, Steckel, Young, Scott and Tranel2017) developed a derived cleaved amplified polymorphic sequences (dCAPs) assay to rapidly identify the presence or absence of R128 PPX2 mutations within Palmer amaranth. R128G and R128M (referred to as R98G and R98M in Giacomini et al. Reference Giacomini, Umphres, Nie, Mueller, Steckel, Young, Scott and Tranel2017) substitutions are conferred by changes at two different nucleotide positions in the PPX2 sequence; therefore, two dCAPS assays were used. Each assay required a nested PCR approach using the AmPPX2LpcF1 (5′-TCCATTACCCACCTTCACC-3′) and AmPPX2LspR1 (5′-TTACGCGGTCTTCTCATCCAT-3′) primers followed by a second amplification using dCAPS primers. The R128M mutation was detected using the dCAPS primers R128-F (5′-CTTGGATACGTGAGAAGCAACAGTTG-3′) and R128-R (5′-TAGCAACGGAAGACCATCTCTATCTAGGTAC-3′). The same forward primer (R128-F) was used in conjunction with an additional reverse primer R128G-R (5′-TAGCAACG-GAAGACCATCTCT ATCTATGAAGC-3′) to detect the R128G mutation. The PCR products were mixed with one unit of the appropriate restriction enzyme (KpnI-HF for R128M and HindIII-HF for R128G, NEB #R3142S and #R3104S) into 1× CutSmart Buffer (New England BioLabs, Inc., Ipswich, MA) and digested overnight (approximately 12 h) at 37 C. Fully, partially, and nondigested products were scored as wild type, heterozygous, and homozygous mutants, respectively.
Results and Discussion
Complete Palmer amaranth control (i.e., 100% mortality) was noted at LC3, OC2, and SC2 field locations (Table 1; Figure 1). PPO-susceptible fields were found in both the North and South region of west Tennessee. In contrast, 15 of the 18 fields tested (83%) had Palmer amaranth survive the fomesafen application. PPO-R Palmer amaranth was found in all regions (North, Central, and South) (Table 1; Figure 1). These observations confirmed widespread resistance to fomesafen throughout west Tennessee.
Genomic DNA of putative PPO-R Palmer amaranth from 15 fields was analyzed to detect whether the ΔG210 resistance mechanism was associated with PPO-R. The ΔG210 mutation was detected in 11 of the 15 fields harboring PPO-R Palmer amaranth, with frequencies ranging from 10% to 70% (Table 2; Figure 2). All individual plants containing the ΔG210 mutation were heterozygous. Of the three known PPX2 mutations, the ΔG210 deletion accounted for 47% of PPO-R Palmer amaranth described in this study (Figure 3). Plants from LC2 and OC1 had only the ΔG210 mutation. In both fields, the ΔG210 mutation was found in 70% of surviving plants (Table 2). However, seven fields (46%) were found to contain both the ΔG210 mutation and R128G mutation in separate PPO-R Palmer amaranth plants (Table 2; Figure 2). These findings are similar to observations in Arkansas, where Varanasi et al. (Reference Varanasi, Brabham, Norsworthy, Nie, Young, Houston, Barber and Scott2017) noted that 27% of accessions tested were segregated and harbored both the ΔG210 mutation and R128G or R128M mutations. The ΔG210 mutation was characterized in 41% of fields within the Central region of west Tennessee (Figures 1, 3, and 4).
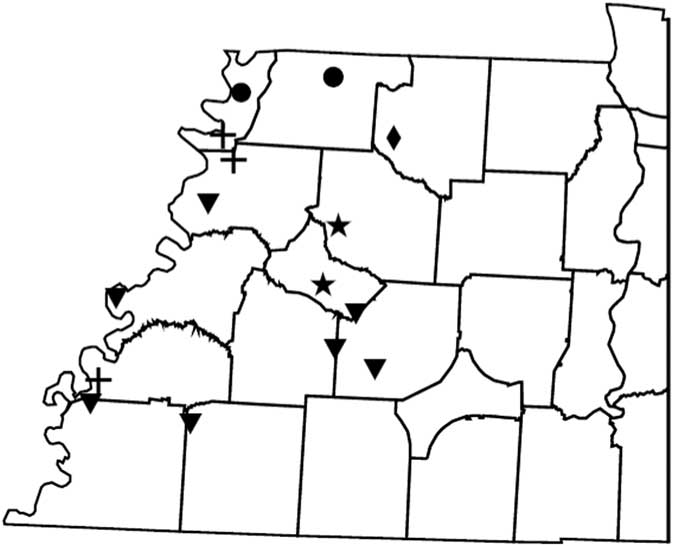
Figure 2 Distribution of PPX2 mutations in Palmer amaranth from west Tennessee. A TaqMan qPCR assay was used to detect the presence of the ΔG210 mutation in the PPX2 gene, and dCAPs assays were used for detection of the R128G and R128M mutations in the PPX2 gene of Palmer amaranth. PPO-resistance mutations: ΔG210 (circles), R128G (diamonds), ΔG210 and R128G (inverted triangles), R128G and R128M (crosses), ΔG210, R128G, and R128M (stars).
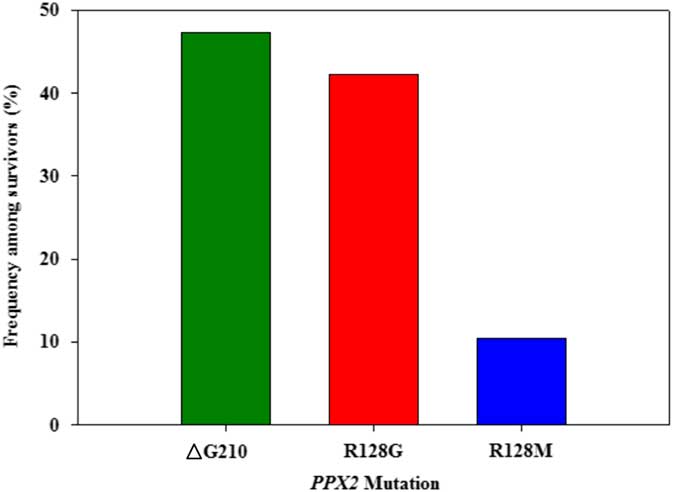
Figure 3 Frequency of each PPX2 mutation among Palmer amaranth plants identified as resistant to fomesafen within west Tennessee. A TaqMan qPCR assay was used to detect the presence of the ΔG210 mutation in the PPX2 gene, and dCAPs assays were used for detection of the R128G and R128M mutations in the PPX2 gene of Palmer amaranth.
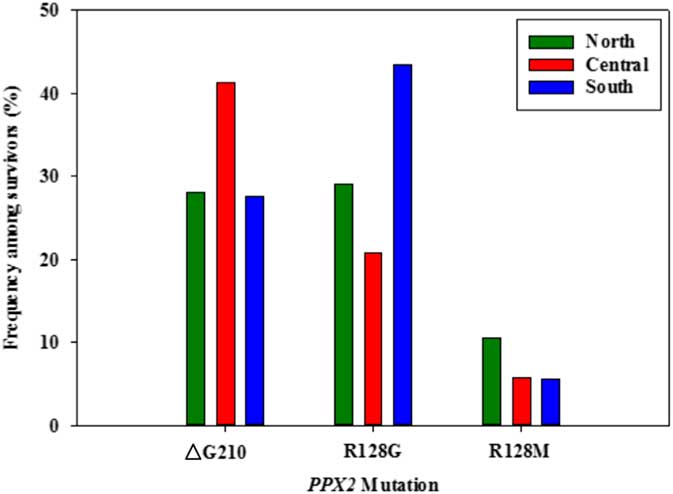
Figure 4 Frequency of each PPX2 mutation among Palmer amaranth plants identified as resistant to fomesafen herbicides within three regions of west Tennessee. A TaqMan qPCR assay was used to detect the presence of the ΔG210 mutation in the PPX2 gene, and dCAPs assays were used for detection of the R128G and R128M mutations in the PPX2 gene of Palmer amaranth.
Table 2 Percentage of the three PPX2 mutations among surviving Palmer amaranth populations of plants with three mutations known to confer resistance to protoporphyrinogen IX oxidase–inhibiting herbicides.
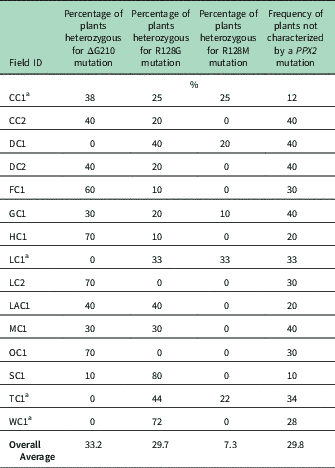
a Number of plants assayed: CC1, eight plants; LC1, nine plants; TC1, nine plants, and WC1, seven plants. At other listed locations, 10 plants were assayed.
The R128G mutation was detected in 13 of the 15 fields tested (Table 2; Figure 2). Much as with the ΔG210 mutation, plants homozygous for R128G were not detected. The frequency of plants heterozygous for the R128G mutation ranged from 10% to 80% in 13 of the 15 fields tested (Table 2). Overall, the R128G mutation accounted for 42% of the PPO-R Palmer amaranth described in this study (Figure 3). In the North and Central region of west Tennessee, the R128G mutation was discovered in 29% and 20% of plants tested, respectively (Figure 4). The R128G mutation was identified in 43% of Palmer amaranth found in the South region of west Tennessee near Memphis (Figures 1, 2, and 4). Likewise, the R128G mutation was identified in 55% of accessions from Crittenden and Lee counties in Arkansas, which are also near Memphis, TN (Varanasi et al. Reference Varanasi, Brabham, Norsworthy, Nie, Young, Houston, Barber and Scott2017). The R128M mutation was discovered in five fields collectively representing all three regions of west Tennessee. (Table 2; Figures 2 and 4). As with the other two mutations, R128M was only found to be heterozygous. The R128M mutation accounted for only 10% of the PPO-R Palmer amaranth described in this study (Figure 3). However, in three fields both the R128G and R128M mutation were found in separate PPO-resistant Palmer amaranth plants (Table 2; Figure 2). Furthermore, at CC1 and GC1, all three known PPX2 mutations (ΔG210, R128G, and R128M) were identified in separate plants at frequencies of 38%, 25%, and 25% and 30%, 20%, and 10%, respectively (Table 2; Figure 2).
Resistance of all surviving Palmer amaranth from each field was not successfully described by the three PPX2 mutations (Table 2; Figure 2). Depending on the field, the frequency of plants not containing one of the three PPX2 mutations ranged from 10% to 40% (Table 2). Similarly, Varanasi et al. (Reference Varanasi, Brabham, Norsworthy, Nie, Young, Houston, Barber and Scott2017) reported that 27 of 167 accessions not controlled by fomesafen contained no known PPX2 mutations. These data indicate the potential for an unknown target-site mutation or metabolic resistance in midsouthern Palmer amaranth populations (Salas-Perez et al Reference Salas-Perez, Burgos, Rangani, Singh, Refatti, Pivetam, Tranel, Mauromoustakos and Scott2017; Varanasi et al. Reference Varanasi, Brabham, Norsworthy, Nie, Young, Houston, Barber and Scott2017). It is interesting that none of the three known mutations was found in the homozygous state. A likely explanation for this is that evolution of resistance to PPO inhibitors is a relatively recent event.
In west Tennessee, 15 of the 18 fields tested harbored Palmer amaranth plants that were not controlled by a POST fomesafen application, indicating that fomesafen resistance is present in these fields. Furthermore, 11 of the 15 fields were characterized by the presence of at least two of the known PPX2 mutations. Schwartz-Lazaro et al. (Reference Schwartz-Lazaro, Norsworthy, Scott and Barber2017) reported that a Palmer amaranth population with both the ΔG210 mutation and R128G mutation had cross-resistance to the five PPO inhibitor chemical families when compared to a single susceptible Palmer amaranth biotype. In this study, researchers conducted a dose-response under greenhouse conditions with five PPO-inhibiting herbicides (flumioxazin, fomesafen, saflufenacil, sulfentrazone, and oxadizon) applied PRE and four PPO-inhibiting herbicides (flumioxazin, fomesafen, saflufenacil, and carfentrazone) applied POST. Complete control was achieved at the 8× rate for PPO-inhibiting herbicides applied PRE and 32× rate for herbicides applied POST (Schwartz-Lazaro et al. Reference Schwartz-Lazaro, Norsworthy, Scott and Barber2017). Results from Schwartz-Lazaro et al. (Reference Schwartz-Lazaro, Norsworthy, Scott and Barber2017) indicate very clear cross-resistance to PPO-inhibiting herbicides applied POST to Palmer amaranth harboring both the ΔG210 and R128G mutations. The results of our study coupled with those from Schwartz-Lazaro et al. (Reference Schwartz-Lazaro, Norsworthy, Scott and Barber2017) would suggest that the fomesafen-resistant Palmer amaranth is also resistant to other PPO-inhibiting herbicides.
However, determining resistance to PRE applications of these herbicides would require further research to verify the findings in a greenhouse setting provided by Schwartz-Lazaro et al. (Reference Schwartz-Lazaro, Norsworthy, Scott and Barber2017). In 2017, field research was conducted to evaluate the effectiveness of PPO-inhibiting herbicides applied PRE on PPO-R and PPO-S Palmer amaranth (Copeland et al. Reference Copeland, Wiggins and Steckel2018). Effective dose values of flumioxazin, sulfentrazone, and saflufenacil for 75% control (ED75) of Palmer amaranth were greater at the PPO-R site compared to the PPO-S site 35 DAT. For instance, ED75 values of flumioxazin at PPO-R site (121 g ai ha–1) were 10 times greater than the PPO-S site (12 g ai ha–1) 35 DAT. However, ED75 values were similar for the aforementioned herbicides at both sites 21 DAT. These findings suggest that PPO-inhibiting herbicides applied PRE have efficacy on PPO-R Palmer amaranth. However, the contributions of the R128G and R128M mutations to PPO-inhibiting herbicides applied PRE and POST are still unknown for Palmer amaranth. Reports from preliminary greenhouse studies have provided that PPO-R waterhemp with the R128G mutation responded similarly to POST applications of fomesafen compared to PPO-R waterhemp with the ΔG210 mutation (Steppig et al. Reference Steppig, Mansfield, Haozhen, Young and Young2017; B. Young, personal communication). Future research should investigate if the PPX2 mutations are affecting Palmer amaranth efficacy of other herbicide families. Moreover, if future research could determine whether all PPX2 mutations provide Palmer amaranth with the same level of resistance to fomesafen applied both PRE and POST, that information could be useful in putting together Palmer amaranth management strategies.
Growers that have fields infested with similar glyphosate and PPO-R Palmer amaranth should use effective herbicide-resistant crops (i.e., glufosinate-, dicamba-, or 2,4-D-resistant crops) with residual herbicides (e.g., chloroacetamides and triazines) that deliver multiple, effective sites of action targeting Amaranthus spp. However, sole reliance on herbicides for a weed management plan is not a sustainable practice (Norsworthy et al. Reference Norsworthy, Ward, Shaw, Llewellyn, Nichols, Webster, Bradley, Frisvold, Powles, Burgos, Witt and Barrett2012). Growers should use integrated weed management strategies to reduce selection pressure for further herbicide resistance. Incorporating cultural practices such as cover crops or narrow row spacing can suppress weeds while reducing the number of herbicide applications in a growing season (Jabran and Chauhan et al. Reference Jabran and Chauhan2018; Wiggins et al. Reference Wiggins, McClure, Hayes and Steckel2016).
Acknowledgments
The Tennessee Soybean Promotion Board provided funding for this research. No conflicts of interest have been declared.










