The giant panda (Ailuropoda melanoleuca), crowned as a national treasure, is an iconic conservation species in China. According to the Fourth National Giant Panda Survey, it is estimated that the wild population consists of about 1,864 animals confined to six fragmented mountain ranges along the eastern edge of Tibet Plateau (State Council Information Office, 2015). Sixty-five giant panda reserves and three large breeding centers have been established for in situ and ex situ conservation, respectively (Wei et al., Reference Wei, Hu, Zhu, Bruford, Zhan and Zhang2012). In captivity, the proportion of twins has accounted for 54% of the total number of births (Wang et al., Reference Wang, Hou, Liu, Cai, Luo, Huang and Zhang2015). Natural discard of a twin by the panda mother resulted in a historic neonatal mortality rate of about 60% (Shan et al., Reference Shan, Hu, Zhu, Yan, Wang, Li and Wei2014). After nearly half a century of efforts, giant panda artificial breeding technology has been successfully applied to rearing the twin generally discarded by panda mothers, and panda twins’ survival rate currently approaches 95% (Wang et al., Reference Wang, Hou, Liu, Cai, Luo, Huang and Zhang2015). By 1997, the number of captive-born giant pandas outnumbered wild-born pandas in the ex situ population (Zhang et al., Reference Zhang, Zhang, Hou, Wang, Li, Fei and Maple2006).
Twins are divided into monozygotic (MZ, ‘identical’) and dizygotic (DZ, ‘fraternal’) twins. DZ twin pairs arise from two fertilization events, while MZ twin pairs most likely arise from splitting of a single early embryo (Cutler et al., Reference Cutler, Murphy, Hopper, Keogh, Dai and Craig2015). Accurate zygosity information of giant panda twin pairs would help managers to improve management of twins’ artificial brood, understand their developmental events, and evaluate their medical life histories. However, research about giant panda zygosity diagnosis has not been carried out, and the proportion of DZ and MZ twins in panda populations remains to be answered.
Comparing the unique genetic fingerprints that can discriminate between individuals offers the most robust method for estimating the zygosity of a twin pair (Forget-Dubois et al., Reference Forget-Dubois, Pérusse, Turecki, Girard, Billette, Rouleau and Tremblay2003; Jackson et al., Reference Jackson, Snieder, Davis and Treiber2001). In this study, we aimed to accurately assess the zygosity of panda twins using short tandem repeat (STR) data. We also aimed to determine the proportion of DZ and MZ twins at the China Conservation and Research Center for the Giant Panda, Wolong (hereafter Wolong), the largest captive breeding center in China. We expect our results to deepen understanding of giant panda breeding and yield insight into the mechanism behind the formation of panda twins.
Materials and Methods
Samples
By the end of 2016, the total captive giant panda population was 469, including 103 pairs of twins. The number of twins thus accounts for 44% of the population. Among panda twins, 45% are pigeon pairs (different-sex), while 55% are same-sex twins.
According to the studbook, there are 45 pairs of panda twins living in the captive population of Wolong (243 pandas in total by the end of 2016): 25 pigeon pairs and 20 same-sex twins. Considering all pigeon pairs are DZ, blood or fecal samples were only collected from 18 pairs of same-sex twins (the missing two pairs were exhibited in other zoos during the study period). All blood samples were obtained from routine health examinations. Total genomic DNA of blood and feces were obtained using Qiagen Dneasy Blood & Tissue Kit and Qiagen QIAamp DNA Stool Mini Kit, respectively, according to the manufacturer's instructions.
PCR Protocol and STR Genotyping
We used 10 tetra-microsatellite loci to distinguish genotype between twin pairs. These were as follows: gpz-47, gpz-6, GPL-47, GPL-60, GPL-29, gpz-20, GPL-53, GPL-44, gpz-51, and GPL-8 (Huang et al., Reference Huang, Li, Du, Yang, Shen, Zhang and Yue2015). PCR amplifications were carried out in 25 μL reaction mixtures, comprising approximately 50 ng of template DNA, 2 mmol MgCl2, 200 μmol of dNTP each, 15 pmol of each primer, 1.0 μg of bovine serum albumin (BSA), 2.5 μL 10 × PCR buffer, and 0.3 units of Hotstart DNA polymerase (Takara). Amplifications were performed using the following PCR procedure: an initial denaturation step for 5 min at 95 °C, followed by 35 cycles of 95 °C for 45 s, 30 s at locus-specific annealing temperature specified in Huang et al. (Reference Huang, Li, Du, Yang, Shen, Zhang and Yue2015) and 50 s at 72 °C, and a final elongation for 10 min at 72 °C. For all 10 markers, the 5´-end of the forward primers was fluorescently labeled. For genotyping, the PCR amplification products were separated by capillary electrophoresis using a denaturing acrylamide gel matrix on an ABI PRISM 377 Genetic Analyzer; 1 μL amplification product and 1 μL formamide loading buffer were mixed with 1 μl GeneScan TAMRA 350 internal size standard (ABI), heated at 95°C for 3 min and flash cooled on ice. Samples were electrophoresed at 15 KV for 2,000 sec. Alleles were detected using the GeneScan/Genotyper software package of Applied Biosystems. The peak amplitude thresholds (PATs) value employed in this study was 200.
All samples were amplified at least three times for each marker. A single-locus genotype was not accepted until our replicates resulted in at least three identical homozygote profiles or two identical heterozygote profiles. These criteria were based on a pilot study, where genotypes obtained from feces versus blood samples were compared (Huang et al., Reference Huang, Li, Du, Yang, Shen, Zhang and Yue2015).
Zygosity Testing
We used MStools for Microsoft Excel to identify matching genotypes in panda twin pairs: (1) If alleles were different at two or more locations, the twin pairs were accepted as DZ. (2) If only one mismatch of one allele was found, DNA was re-extracted and three more PCR replications were performed. If it was still different, we judged the twin pairs as DZ. (3) If all alleles in all loci were identical, the twin pairs were believed to be MZ. We then calculated the average non-exclusion probability of identity among siblings across these 10 microsatellite loci in the captive population of Wolong (n = 113) using CERVUS 3.0 (Marshall et al., Reference Marshall, Slate, Kruuk and Pemberton1998). We conducted tests for Hardy–Weinberg equilibrium (HWE) and linkage disequilibrium (LD) on a subset of the Wolong captive population with available genetics data (n = 113). CERVUS 3.0 was used to test HWE. The LD test was done in GENEPOP V4.2 (Raymond & Rousset, Reference Raymond and Rousset1995; Rousset, Reference Rousset2008).
Results
The tests for HWE showed significant deviations from HWE at loci GPL-8 and gpz-47 (p < .05) in the Wolong captive population. The other 8 loci did not deviate significantly from HWE and all 10 grouped loci did not significantly deviate from HWE after Bonferroni correction (adjusted p = .05/10; see Table S1). Of the 10 microsatellite loci out of the 45 possible microsatellite locus pairs, 5 pairs — GPL-60/gpz-20, GPL-60/GPL-53, GPL-53/gpz-47, GPL-47/gpz-20, and GPL-29/GPL-8 — were in significant LD after applying Bonferroni corrections (adjusted p = .05/10; see Table S2).
The probability of two individuals sharing an identical multi-locus genotype was 0.00037, based on 10 loci, indicating that this subset of 10 loci was enough for accurate individual identification (PIDsib < 0.01) in our target population (Waits et al., Reference Waits, Luikart and Taberlet2001). The probability of monozygotic twins was 99.963% when all 10 markers were concordant.
Zygosity diagnosis was performed through comparing the concordance of the twin-pair genotypes at 10 microsatellite markers (Figure 1 about here). Of the 18 studied same-sex twin pairs in Wolong, discordant loci were found in every pair, which indicates that all 18 twin pairs are DZ twins (Table 1 about here). When considering the 25 pairs of pigeon twins in addition to the 18 pairs of same-sex twins, no MZ twins were found in the 43 total pairs in this study.
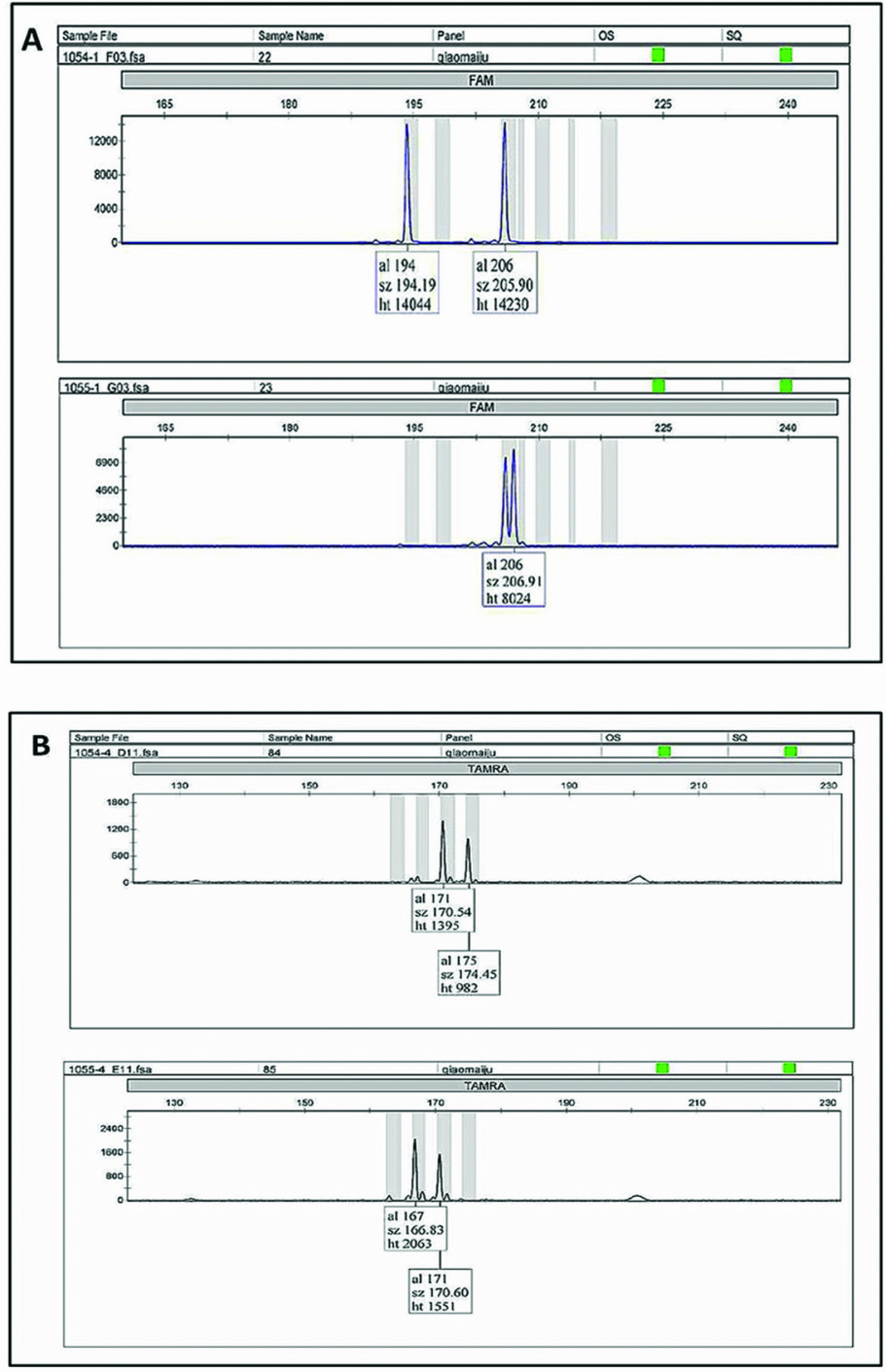
FIGURE 1 Microsatellite loci genotypes: A and B indicate the genotype of male twin pairs ‘Qin Qin’ (stud number 1054) and ‘Ai Ai’ (stud number 1055) at locus gpz-6 and GPL-29, respectively. Because both alleles were different, the twin pairs were accepted as DZ.
TABLE 1 The Genotype and Zygosity of 18 Same-Sex Giant Panda Twin Pairs

Note: 0 represents null alleles.
Discussion
Previous twin zygosity studies of humans have calculated the probability of any twin pair to be MZ at five concordant microsatellite markers to be 99% (Becker et al., Reference Becker, Busjahn, Faulhaber, Bähring, Robertson, Schuster and Luft1997), and up to 99.9% with nine concordant markers (He et al., Reference He, Zhu, Han, Liu, Wang, Chu and Huang2001). Our twin zygosity analysis of giant pandas was performed with 10 tetra-microsatellite markers, and the probability of being MZ was 99.963% if all 10 markers were concordant. Our method is thus a rapid and accurate approach to twin zygosity determination in giant pandas.
Our giant panda twin zygosity diagnosis revealed that all 43 twin pairs were DZ, while no MZ twins were diagnosed. The proportion of MZ and DZ twins in humans varies within different populations, with close to 1:1 in Japan, about 1:5 in some parts of India, and 1:7 in central African countries (Bortolus et al., Reference Bortolus, Parazzini, Chatenoud, Benzi, Bianchi and Marini1999; Derom et al., Reference Derom, Orlebeke, Eriksson, Thiery, Keith, Papiernik, Keith and Luke1995; Oleszczuk et al., Reference Oleszczuk, Keith, Keith and Rayburn1999; Smits & Monden, Reference Smits and Monden2011), while our result for panda twins in this study was 0:43. We speculate that the fertilized eggs of giant panda do not have the capability to split into two identical embryos, or that this capability is very poor. This is likely due to the delayed implantation of giant panda embryos (Sutherland-Smith et al., Reference Sutherland-Smith, Morris and Silverman2004; Zhang et al., Reference Zhang, Li, Wang and Hull2009), in which the embryo floats in the womb and development is arrested for about 3 months. The best time for fertilized eggs to split and form two identical embryos is thus probably missed. Once the fetus is implanted in the uterus wall, it takes just 15–20 days before the underdeveloped panda cubs are born.
STR cannot distinguish MZ twins, because they are genetically nearly identical. Considering twins account for 54% of the total number of births, researchers have historically worried that identical twins could be misjudged as one individual, resulting in underestimated population sizes in wild giant panda surveys. Based on our results, there are either no identical panda twins or the incidence is very low, and coupled with the tendency of panda mothers to abandon one twin, we conclude that there is no impact of MZ twins on the accuracy of wild giant panda population size estimation. Because there was some evidence that the captive panda population at Wolong likely has skewed allele frequencies due to deviation from HWE at and LD between some loci and locus pairs, respectively, our calculated probability of monozygosity at 10 concordant loci is likely an overestimate. That said, although the allele frequencies between captive pandas and wild populations are likely very different, the somewhat inbred captive population in fact strengthens our conclusion that MZ twins are absent or extremely low in giant pandas. This is because we would expect more DZ twins to potentially appear as MZ due to the loss of alleles in the captive population and higher incidence of repeated genotypes by chance. Because we still found no MZ twins, we can conclude with more certainty that MZ twins have little to no effect on surveys of wild giant panda populations. Twin studies, such as that conducted here, can have important applications to both in situ and ex situ wildlife conservation and thus should be emphasized more in the future.
Acknowledgment
We thank all the panda breeders for their help with the collection of fecal samples. This work was supported by the central finance forestry national nature reserve subsidy fund — Sichuan Wolong National Nature Reserve (grant number: Sichuan Forestry 2016–923) and a special grant from the Giant Panda International Cooperation Research Project (grant number: State Forestry Administration of the People's Republic of China 2017–115).
Disclosure of Interests
None.
Details of Ethical Approval
The authors assert that all procedures contributing to this work comply with ethical standards of the relevant national and institutional guides on the care and use of laboratory animals.
Supplementary material
To view supplementary material for this article, please visit https://doi.org/10.1017/thg.2018.59




