Neurodevelopmental disorders (NDDs), such as autism spectrum disorder (ASD) and attention-deficit hyperactivity disorder (ADHD), are a group of early onset conditions affecting approximately 10–15% of the population (Boyle et al., Reference Boyle, Boulet, Schieve, Cohen, Blumberg, Yeargin-Allsopp and Kogan2011). NDDs are characterized by alterations in developmental trajectories causing impairments in personal, social, academic, and occupational functioning (American Psychiatric Association, 2013). There is substantial co-occurrence of different NDDs, suggesting a common etiology for the disorders (Thapar et al., Reference Thapar, Cooper and Rutter2017). NDDs have a strong genetic component, and the familial risk is seen between different NDDs and psychiatric disorders (Faraone et al., Reference Faraone, Ghirardi, Kuja-Halkola, Lichtenstein and Larsson2017; Ghirardi et al., Reference Ghirardi, Brikell, Kuja-Halkola, Freitag, Franke, Asherson and Larsson2017; Sullivan et al., Reference Sullivan, Magnusson, Reichenberg, Boman, Dalman, Davidson and Lichtenstein2012). The etiological overlap has also been observed in molecular genetic studies. Many of the same risk genes and variants, identified through genome analyses of copy number variants (CNVs) and sequence level variants are found across NDDs (Torres et al., Reference Torres, Barbosa and Maciel2016). These include large deletions and duplications of 16p11.2 and 22q11.2, duplication of 7q11.23 and 15q11-13 as well as smaller CNVs affecting specific genes such as NRXN1 (OMIM#600565) and SHANKs (Kirov, Reference Kirov2015). Additionally, new NDD risk genes have been implicated using sequencing-based studies, mainly through identification of de novo mutations (DNMs) (De Rubeis et al., Reference De Rubeis, He, Goldberg, Poultney, Samocha, Cicek and Buxbaum2014; Iossifov et al., Reference Iossifov, O'Roak, Sanders, Ronemus, Krumm, Levy and Wigler2014; Tammimies et al., Reference Tammimies, Marshall, Walker, Kaur, Thiruvahindrapuram, Lionel and Fernandez2015; Yuen et al., Reference Yuen, Merico, Bookman, Howe, Thiruvahindrapuram, Patel and Scherer2017).
In addition to germline DNMs, somatic DNMs contribute to human disorders (Acuna-Hidalgo et al., Reference Acuna-Hidalgo, Veltman and Hoischen2016), including NDDs (Lim et al., Reference Lim, Uddin, De Rubeis, Chan, Kamumbu, Zhang and Walsh2017; Yuen et al., Reference Yuen, Merico, Cao, Pellecchia, Alipanahi, Thiruvahindrapuram and Scherer2016). It has been hypothesized that postzygotic DNMs could cause discordant phenotypes in monozygotic (MZ) twin pairs that are otherwise genetically identical. However, the majority of studies testing this hypothesis have reported no or limited evidence for the postzygotic de novo CNVs in MZ discordant twin pairs for psychiatric disorders (Bloom et al., Reference Bloom, Kahler, Collins, Chen, Cannon, Hultman and Sullivan2013; Castellani et al., Reference Castellani, Awamleh, Melka, O'Reilly and Singh2014) and neurodevelopmental problems (Abdellaoui et al., Reference Abdellaoui, Ehli, Hottenga, Weber, Mbarek, Willemsen and Boomsma2015; Ehli et al., Reference Ehli, Abdellaoui, Hu, Hottenga, Kattenberg, van Beijsterveldt and Davies2012; Laplana et al., Reference Laplana, Royo, Aluja, Lopez, Heine-Sunyer and Fibla2014). In rare occasions, postzygotic CNVs can also be found in MZ pairs without discordant phenotypes (Abdellaoui et al., Reference Abdellaoui, Ehli, Hottenga, Weber, Mbarek, Willemsen and Boomsma2015; Magnusson et al., Reference Magnusson, Lee, Chen, Szatkiewicz, Pramana, Teo and Pawitan2016; McRae et al., Reference McRae, Visscher, Montgomery and Martin2015).
Here, we analyze the presence of postzygotic CNVs in a Swedish twin sample from the Roots of Autism and ADHD Study in Sweden (RATSS; Bölte et al., Reference Bölte, Willfors, Berggren, Norberg, Poltrago, Mevel and Lichtenstein2014). The sample is enriched for twin pairs discordant for NDDs, including 13 MZ twin pairs discordant for ASD. Beyond rare case reports (Laplana et al., Reference Laplana, Royo, Aluja, Lopez, Heine-Sunyer and Fibla2014), there have not been studies focused on ASD-discordant MZ pairs and the contribution of postzygotic CNVs. Additionally, we characterized the rate of shared rare CNVs in genes implicated earlier in brain development among the twin pairs by grouping them as concordant or discordant for NDD and ASD and comparing with typically developing (TD) twin pairs.
Materials and Methods
Participants and Clinical Assessments
All the participants included in this study are part of the RATSS (Bölte et al., Reference Bölte, Willfors, Berggren, Norberg, Poltrago, Mevel and Lichtenstein2014). The recruitment and procedure for the sample collection have been described in detail elsewhere (Bölte et al., Reference Bölte, Willfors, Berggren, Norberg, Poltrago, Mevel and Lichtenstein2014; Willfors et al., Reference Willfors, Carlsson, Anderlid, Nordgren, Kostrzewa, Berggren and Bölte2017). Overview of the study design and samples used for the analyses are shown in Figure 1. Within the RATSS, twins undergo detailed diagnostic and psychological assessments, including standardized diagnostic tools for NDDs: a psychosocial and anamnestic interview, the Autism Diagnostic Interview-Revised (ADI-R), the Autism Diagnostic Observation Schedule Second Edition (ADOS-2), the Kiddie Schedule for Affective Disorders and Schizophrenia (K-SADS), or the Diagnostic Interview for ADHD in Adults (DIVA). Additionally, the Wechsler Intelligence Scales for Children or Adults, Fourth Edition (WISC-IV) or the Leiter-revised scales in combination with the Peabody Picture Vocabulary Test, Third Edition, and the parent-rated Adaptive Behavior Assessment Scale, 2nd Edition (ABAS-2), were used to evaluate adaptive, cognitive, and verbal abilities and for diagnostic assessments. Also, a medical and family history of the twins was obtained by parental interviews and by collecting existing medical records.

FIGURE 1 Overview of the study including the numbers of monozygotic (MZ) and dizygotic (DZ) pairs included in each step of the analysis. The dashed arrows indicate samples and copy number variants (CNVs) were excluded from the analysis. *Three twin pairs were excluded as psychiatric disorders were present in the pairs.
The given NDD diagnoses are based on the DSM-5 criteria (American Psychiatric Association, 2013) by incorporating the different assessments followed by a consensus among three experienced clinicians within the RATSS study. Using the Child and Adolescent Twin Study in Sweden cohort, which is also a recruitment cohort for RATSS, it has been demonstrated that in the majority of ASD-discordant MZ pairs, the co-twin has another NDD diagnosis (Lundstrom et al., Reference Lundstrom, Reichenberg, Melke, Rastam, Kerekes, Lichtenstein and Anckarsater2015). Therefore, in this study, we used two strategies to categorize the twins, based on their NDD diagnoses. First, we only considered whether any of the twins had ASD diagnosis and categorized the pair as discordant or concordant for ASD. Thereafter, we considered all NDD diagnoses and did an additional categorization as NDD discordant (affected twin has any NDD diagnosis and co-twin TD) and NDD concordant (both twins have at least one NDD diagnosis). If no NDD diagnoses were present in any of the twins, the pair was categorized as TD.
Figure 1 shows the overview of the study details and number of samples within our analysis. A total of 208 twins from 104 twin pairs (a triplet was counted as one MZ female pair and one dizygotic (DZ) brother) were included, collected within RATSS between 2011 and 2014. In three families, samples from two twin pairs were included. Additionally, 75 parents from 37 of these families were analyzed. After the quality control of genotyping and CNV data, the sample set used for the CNV analysis comprised of 69 MZ (including one female MZ pair from the triplet) and 31 DZ twin pairs (the male sibling from the triplet was excluded from the analysis), corresponding to 97 families. A total of 26 pairs were discordant and 11 pairs concordant for ASD (Table 1). In the twin pairs discordant for ASD, 69.2% (18/26) of the affected twins and 19.2% (5/26) of the co-twins had at least one additional NDD diagnosis. For the NDD category, 34 pairs were classified as discordant and 35 pairs as concordant. Twenty-seven twin pairs were classified as typically developed. Mean age, IQ, and the number of other DSM-5 NDD diagnoses for the twin pairs in each category are shown in Table 1.
TABLE 1 Phenotypic Characteristics of 66 Monozygotic and 30 Dizygotic Twins
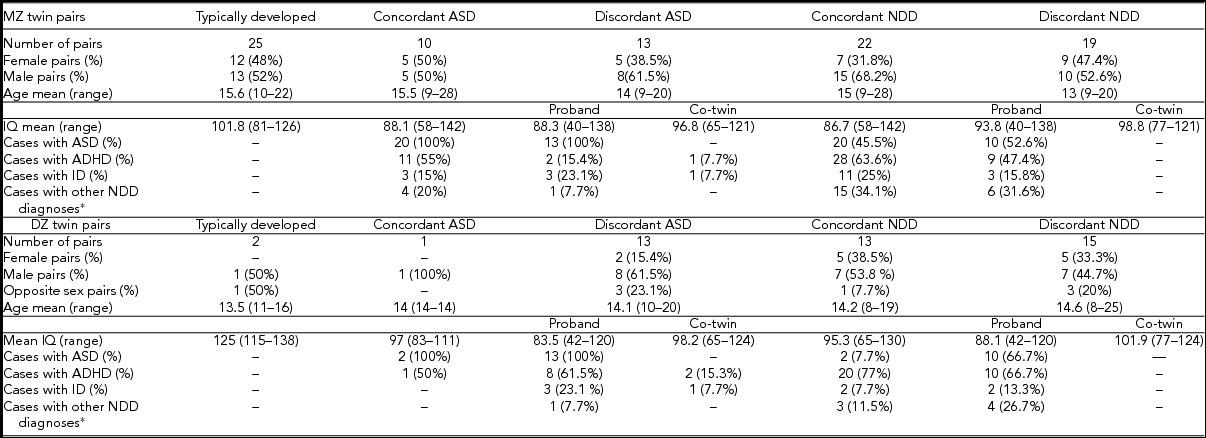
*Additional NDD diagnosis include specific learning disorder, tic disorder, and speech sound disorder.
DNA Samples and Genotyping
Saliva samples were collected from the twins and their parents using the Oragene•DNA OG-500 tubes (DNA Genotek, Inc., Ottawa, Ontario, Canada) during the RATSS study visit or at home. The extraction of genomic DNA from the saliva samples was done automatically using the Autopure LS system and Puregene DNA purification kit (Gentra Systems, Minneapolis, MN, USA) or Chemagen DNA saliva kit (PerkinElmer, Waltham MA, USA). A total of 200 ng of DNA was used for the genotyping using the Infinium PsychArray-24 v1.1 (Illumina Inc., San Diego, California, USA), which contains a total of ~593,260 SNP markers, including 50,000 specific genetic markers for neuropsychiatric disorders, according to the manufacturer's protocol. Samples with SNP call rate lower than 98% were excluded from further investigations. The zygosity of the twin pairs was confirmed using the genotype data by estimating the identity by descent using the PLINK/1.07 (Purcell et al., Reference Purcell, Neale, Todd-Brown, Thomas, Ferreira, Bender and Sham2007) after quality control and removal of SNPs with minor allele frequency <0.05 within the samples.
CNV Calling
We derived the CNVs in our data by using four different algorithms: PennCNV (Wang et al., Reference Wang, Li, Hadley, Liu, Glessner, Grant and Bucan2007), QuantiSNP (Colella et al., Reference Colella, Yau, Taylor, Mirza, Butler, Clouston and Ragoussis2007), iPattern (Pinto et al., Reference Pinto, Pagnamenta, Klei, Anney, Merico, Regan and Betancur2010), and iPsychCNV (prerelease beta v1.0; Bertalan et al., Reference Bertalan, Weinsheimer, Sparsø, Mazin and Werge2015). Although there is no established procedure for CNV calling using the intensity data from Infinium PsychArray, the combination of multiple CNV detection algorithms is a ‘gold standard’ approach to detect reliable CNV calls and has been widely used with other Illumina arrays (Pinto et al., Reference Pinto, Delaby, Merico, Barbosa, Merikangas, Klei and Scherer2014). The called CNVs were annotated based on GRCh37/hg19 genome build. For the downstream analyses, we used only stringent CNVs, which were called by iPattern, and at least one additional algorithm or by PennCNV together with iPattern or/and iPsychCNV with 50% reciprocal overlap between the algorithms. We further excluded variants that were <10 kb in size, covered by <10 consecutive probes and had more than 50% overlap with centromeres or telomeres. After obtaining the final set of stringent CNV calls, we removed individuals with excessive CNV calls defined by >2 standard deviations from the mean CNV count in the whole sample. Out of the 283 individuals genotyped, a total 231 calls proceeded for further analysis after the QC of genotyping and CNV calling (Figure 1). The majority of the samples that failed QC were parental saliva samples collected at home. For 55 MZ and 19 DZ pairs, both twins were successfully genotyped. In 18 families both parents were genotyped.
Population-based control data sets described by Lionel et al. (Reference Lionel, Tammimies, Vaags, Rosenfeld, Ahn, Merico and Scherer2014) were used to restrict the analyses to rare CNVs found in frequency <0.01% in controls. Additionally, we computed the frequency of CNV events using PLINK/1.07 (Purcell et al., Reference Purcell, Neale, Todd-Brown, Thomas, Ferreira, Bender and Sham2007) based on two reference sets: the total cohort including both parents and the twins, and the parents only. We removed variants present in >3 copies in the parents.
Pre-Twinning and Post-Twinning De Novo CNV Detection
The pre-twinning de novo CNVs were analyzed in 18 families in which DNA was available for both parents. The rare stringent CNVs shared by a twin pair or present in one twin in the twin pairs and not found in the parents or the whole population stringent and non-stringent CNV calls were considered as putative pre-twinning de novo variants. Similarly, the postzygotic de novo CNV events were analyzed using the stringent CNV set from the 55 complete MZ twin pairs. Rare stringent CNVs found only in one twin were further filtered using the population frequencies as well as using the non-stringent calls in the co-twins/families. All the surviving unique CNVs that were not detected in any of the non-stringent calls in the co-twin or the whole population dataset were considered as post-twinning de novo variants. The putative pre- and post-twinning de novo CNV calls were visually inspected by plotting the Log R Ratio (LRR) and B allele frequency values of the twins and the parents. If both LRR and B allele frequency showed a clear pattern of the called copy number (CN) and no deviation from normal CN were found in the other twin or parents, the CNV proceeded for experimental validation.
Rare CNVs Overlapping Genes Implicated in Neurodevelopmental Disorders
Next, we characterized rare CNVs for the MZ and DZ twins groups based on the concordance/discordance for ASD and NDD. We included all MZ twin pairs for which at least one twin was successfully genotyped and each twin in DZ pairs. Among the all rare CNVs, we identified CNVs that overlapped known genomic disorders, earlier implicated in ASD/intellectual disability genes (Pinto et al., Reference Pinto, Delaby, Merico, Barbosa, Merikangas, Klei and Scherer2014), genes curated in the SFARI gene database (accession date September 2016; Banerjee-Basu & Packer, Reference Banerjee-Basu and Packer2010) and genes associated with abnormality of the nervous system in the human phenotype ontology (HP:0000707) hereafter termed as HPO-NS (Groza et al., Reference Groza, Kohler, Moldenhauer, Vasilevsky, Baynam, Zemojtel and Robinson2015). Thereafter, the CNVs were also evaluated based on the criteria provided by the American College of Medical Genetics and Genomics (ACMG; Kearney et al., Reference Kearney, Thorland, Brown, Quintero-Rivera and South2011). Three group Fisher's exact test (FET) were used to test whether any significant changes among the MZ twin pairs within different discordancy groups. McNemar's test was used among the DZ discordant pairs to test for any intra-pair significant differences. All statistical analyses were performed with R (3.2.3).
Experimental Validation of Selected CNVs
Putative pre- and post-zygotic de novo and NDD associated CNVs that passed our filtering criteria and visual inspection were subjected to experimental validation using quantitative PCR (qPCR). For this, Taqman® probes (Thermo Fisher Scientific, Waltham MA, USA) within the genomic region of interest for the validation experiment were chosen and tested together with a reference control probe targeting the TERT gene (OMIM#187270; Thermo Fisher Scientific). Also, the CN state of each region was tested in the twin pair, parental DNA if available, and at least one unrelated control DNA sample. Quadruplicates of each sample were analyzed on StepOne™ (Thermo Fisher Scientific) system according to manufacturer's recommendations. The CN of the target in each sample was analyzed using the CopyCaller™ software (Thermo Fisher Scientific).
Results
Pre-Twinning and Post-Twinning De Novo CNVs
No validated pre-twinning de novo CNVs were found in the 18 families in which both parents were successfully genotyped (Table S1). As our next step, we aimed to identify putative postzygotic de novo CNV events in the 55 complete MZ twin pairs, of which 11 pairs were classified as discordant for ASD or 18 pairs as discordant for NDDs. First, we calculated the concordance rate of each CNV detection algorithm when using the <10 kb size and <10 consecutive probe filter within the twin pairs (Table S2). The average concordance rates ranged between 13.6% for iPsychCNV and 65.4% for iPattern. In our final stringent CNV calls, the concordance rate was 64.2% with 16 of 55 twin pairs (29.1%) sharing all their CNVs.
For CNVs larger than 100 Kb the mean concordance rate among MZ twins was 73.6% (median 84.2%) with 24 of 55 twin pairs (43.6%) sharing all their CNVs consistent with the prior literature (Abdellaoui et al., Reference Abdellaoui, Ehli, Hottenga, Weber, Mbarek, Willemsen and Boomsma2015). After removing the common variants, variants called by lower stringency in the co-twin or present in additional individuals in our dataset (see Materials and Methods, Figure 1), a total of eight non-shared rare stringent putative postzygotic CNV calls were found in four MZ twin pairs in our analysis. Following a visual inspection, five of them proceeded to qPCR validation. None of the five CNVs were validated to be non-shared; two were found in both twins, and three CNVs were false positives (see Table S1).
Rate of Rare CNVs Affecting Genes Previously Implicated in NDDs or Nervous System Disorders
In total, 28 CNVs in 22 twin pairs were found to overlap the pre-defined gene sets, including ASD/ID genes (Pinto et al., Reference Pinto, Delaby, Merico, Barbosa, Merikangas, Klei and Scherer2014) in combination with SFARI genes (Banerjee-Basu & Packer, Reference Banerjee-Basu and Packer2010) and/or HPO-NS genes. From these 28 CNVs, nine were targeted for experimental validation using qPCR (Additional file 2, Supplementary Table S3) and all of these CNVs were validated. We calculated the proportion of twin pairs or twins in discordant DZ pairs having at least one CNV in these gene categories within each twin category. We found an increased rate of MZ twin pairs discordant and concordant for ASD having CNVs affecting both NDD genes (Figure 2A) and HPO-NS (Figure 2B) in comparison with TD MZ pairs. However, only the HPO-NS comparison reached statistical significance (three-group comparison 2-sided FET p = .02). As expected, the largest difference was found between the frequency in TD pairs compared with concordant ASD pairs (OR = 14.4, 95% CI [1.2, 814.6]). For MZ twins categorized based on the NDD diagnosis, we did not observe any significant differences for the NDD or HPO-NS (three-group comparison, 2-sided FET p = .92, and p = .11, respectively). To note, the proportion of twin pairs with CNVs for both NDD concordant and discordant pairs was higher than in the TD pairs for the HPO-NS genes although not significant (Figure 2D). For DZ twin pairs, no differences were found between ASD/NDD affected twins and their co-twins (Supplementary Table S4).
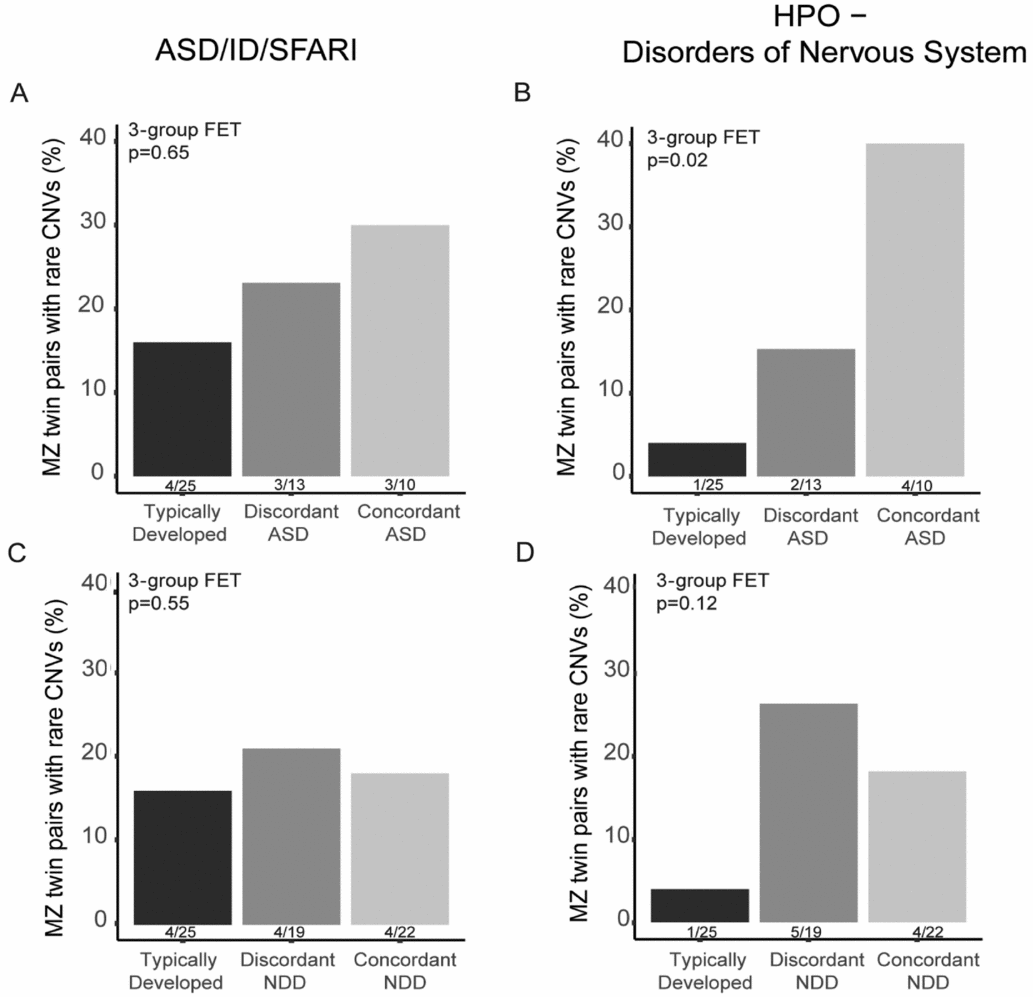
FIGURE 2 The rate of rare CNVs affecting genes earlier implicated in neurodevelopmental disorders (ASD/ID/SFARI) or abnormality of the nervous system in the Human Phenotype Ontology database (HPO-NS) in MZ twin pairs. The percentage of MZ twins in typically developing, ASD-discordant and -concordant pairs with CNVs overlapping genes in ASD/ID/SFARI gene lists (A) and in HPO-NS (B). The percentage of typically developing, NDD-discordant and -concordant pairs with CNVs overlapping genes in ASD/ID/SFARI gene lists (C) and in HPO-NS (D). Statistical significance was tested between the three twin pair groups using Fisher Exact Test, p-value for three group comparison is reported in the figure.
Rare CNVs with Putative Clinical Significance Found in the Sample
Next, we investigated the rare CNVs overlapping the ASD/ID and HPO-NS gene lists to categorize them based on the guidelines from the ACMG (Kearney et al., Reference Kearney, Thorland, Brown, Quintero-Rivera and South2011) and evidence from the earlier literature. Two CNVs in two twin pairs among the 10 MZ pairs concordant for ASD (20.0%) and one additional CNV among the other MZ pairs concordant for NDD (13.6%, 3 out of 22 pairs), were classified as clinically significant. A male MZ twin pair concordant for ASD and ADHD (Family ID 12) had a paternally inherited ~9.0 Mb duplication at 12q11-12q13.11 (GRCh37/hg19, chr12:37,995,364-47,072,741) affecting 33 genes. Proximal deletions of 12q12 had earlier been implicated in patients with NDDs and dysmorphic features (Adam et al., Reference Adam, Mehta, Knight, Hall and Rossi2010). Another MZ male pair (Family ID 23) concordant for ASD and ID and discordant for tic disorder had a ~44.3 kb deletion on chromosome 1q44 (GRCh37/hg19, chr1:244,997,953-245,042,312) affecting COX20 (OMIM#614698) and HNRNPU (OMIM#602869), as well as HNRNPU-AS1 long non-coding antisense RNA. We were not able to conclude whether the deletion was of de novo origin as paternal DNA sample was not available. Recently, multiple reports have described cases with mutations affecting HNRNPU demonstrating its role in NDDs, including severe intellectual disability and early onset seizures (Bramswig et al., Reference Bramswig, Ludecke, Hamdan, Altmuller, Beleggia, Elcioglu and Wieczorek2017; Leduc et al., Reference Leduc, Chao, Qu, Walkiewicz, Xiao, Magoulas and Yang2017; Yates et al., Reference Yates, Vasudevan, Chandler, Donnelly, Stark, Sadedin and Balasubramanian2017). Additionally, a maternally inherited 18.8 kb deletion on chr8p23.2 (GRCh37/hg19, chr8:3,063,043-3,081,903) disrupting CSMD1 (OMIM#608397) was detected in a MZ male pair (Family ID 11) concordant for ADHD and discordant for tic disorder. Recently, both inherited and de novo variants in CSMD1 have been found in ASD patients (Cukier et al., Reference Cukier, Dueker, Slifer, Lee, Whitehead, Lalanne and Pericak-Vance2014; Krumm et al., Reference Krumm, Turner, Baker, Vives, Mohajeri, Witherspoon and Eichler2015). Krumm et al. (Reference Krumm, Turner, Baker, Vives, Mohajeri, Witherspoon and Eichler2015) demonstrated a depletion of CNVs in controls within the 3ʹ genomic loci of CSMD1 and overlap of genomic variants in cases. The deletion detected in the twins is within the region the study highlighted.
Among the discordant MZ pairs, we detected a maternally inherited 0.95 Mb gain on chromosome 6q13-14.1 (GRCh37/hg19, chr6:75,682,566-76,635,743) possibly contributing to the phenotypes seen in the male twin pair (Family ID 29). Both large deletions and duplications at chromosome 6q encompassing this region have been implicated in NDD associated phenotypes (Kaminsky et al., Reference Kaminsky, Kaul, Paschall, Church, Bunke, Kunig and Martin2011; Van Esch et al., Reference Van Esch, Rosser, Janssens, Van Ingelghem, Loeys and Menten2010), and no similar size overlapping CNVs have been detected in DGV. The affected twin has ASD with full-scale IQ of 70, dyslexia, and earlier indicated mild macrocephaly (current head circumference 54.5 cm, 84th percentile). The co-twin has mild macrocephaly (56 cm, 98th percentile) and a reported mild language delay described as language impairments specific for ASD, that is, reversal use of pronouns and regular use of idiosyncratic language, as well as mild understanding and articulation problems. Lack of social reciprocity, pretend, and fantasy play is reported for both twins. However, the impairments in the co-twin are overall reported as previous during development and at a sub-threshold level, and were not observed at the time of the assessment, in comparison to the affected twin. The co-twin also has a slightly higher IQ of 77.
In the DZ pairs, we identified a CNV affecting SYNE1 (OMIM#608441) that is likely pathogenic in a male twin diagnosed with ASD and ID from an opposite-sex DZ pair (Family ID 31). We verified the 39 kb gain on chr6q25.2 (GRCh37/hg19, chr6:152,712,302-152,751,762) using qPCR and showed it to be de novo (Figure S1). His TD female sibling, mother, and father (DNA sample obtained for qPCR validation) had normal CN for the region. SYNE1, which encodes for the Nesprin-1 protein, has been implicated in ASD through rare mutations (De Rubeis et al., Reference De Rubeis, He, Goldberg, Poultney, Samocha, Cicek and Buxbaum2014; Jiang et al., Reference Jiang, Yuen, Jin, Wang, Chen, Wu and Scherer2013; Krumm et al., Reference Krumm, Turner, Baker, Vives, Mohajeri, Witherspoon and Eichler2015; O'Roak et al., Reference O'Roak, Deriziotis, Lee, Vives, Schwartz, Girirajan and Eichler2011) but is also involved in spinocerebellar ataxia (Wiethoff et al., Reference Wiethoff, Hersheson, Bettencourt, Wood and Houlden2016) and Emery–Dreifuss muscular dystrophy (Zhang et al., Reference Zhang, Bethmann, Worth, Davies, Wasner, Feuer and Shanahan2007).
Discussion
We aimed to investigate the presence of postzygotic de novo CNVs that could explain the discordance of a variety of NDD phenotypes, especially ASD, in MZ pairs by genotyping saliva-derived DNA using the Illumina Infinium PsychArray that was designed for studies of psychiatric disorders. We did not identify any validated postzygotically derived unique CNVs in the 55 MZ pairs analyzed, thus indicating that these do not contribute to the discordancy of NDDs in our sample. Our results are in accordance with the earlier studies showing that postzygotic de novo CNVs are rare when analyzing genomic DNA from blood or saliva samples (Abdellaoui et al., Reference Abdellaoui, Ehli, Hottenga, Weber, Mbarek, Willemsen and Boomsma2015; Bloom et al., Reference Bloom, Kahler, Collins, Chen, Cannon, Hultman and Sullivan2013; Ehli et al., Reference Ehli, Abdellaoui, Hu, Hottenga, Kattenberg, van Beijsterveldt and Davies2012; Laplana et al., Reference Laplana, Royo, Aluja, Lopez, Heine-Sunyer and Fibla2014; Magnusson et al., Reference Magnusson, Lee, Chen, Szatkiewicz, Pramana, Teo and Pawitan2016). Earlier literature suggests that a lower level of mosaicism is masked in blood-derived DNA (Erlich, Reference Erlich2011; King et al., Reference King, Sifrim, Fitzgerald, Rahbari, Hobson, Homfray and Hurles2017). Here, we restricted our analysis to saliva DNA and CNV level variants. However, we cannot exclude the possibility that other types or tissue-specific postzygotic somatic variations contribute to the discordancy. In non-twin cohorts, somatic sequence level mutations are found in 7.5% of affected children and shown to contribute to both ASD and ID diagnoses (Acuna-Hidalgo et al., Reference Acuna-Hidalgo, Bo, Kwint, van de Vorst, Pinelli, Veltman and Gilissen2015; Freed & Pevsner, Reference Freed and Pevsner2016; Lim et al., Reference Lim, Uddin, De Rubeis, Chan, Kamumbu, Zhang and Walsh2017). There is no clear estimate of the rate of somatic CNVs, and although our sample is enriched for discordant twins that in theory should have a higher rate of such events, one obvious limitation of our study is the small sample size. Further studies investigating the rate of postzygotic CNV events in discordant twins should be done in larger twin cohorts with detailed phenotype information.
Additionally, somatic CNVs and sequence level variation can be found in human neurons from healthy individuals using single-cell genomics (Lodato et al., Reference Lodato, Woodworth, Lee, Evrony, Mehta, Karger and Walsh2015; McConnell et al., Reference McConnell, Lindberg, Brennand, Piper, Voet, Cowing-Zitron and Gage2013). Therefore, stochastic genetic variations at a neuronal level could contribute to disrupted brain development and underlie phenotype differences connected to brain disorders in MZ twins. As recent literature has implicated the role of somatic DNMs in NDDs, a better understanding of their contribution and origin in NDDs is needed as it can inform better estimations of the recurrence rates of the disorders and thus genetic counseling.
We also sought to understand the contribution of shared rare CNVs in the twin pairs with a specific focus on the ASD and NDD-discordant twins. As expected, we identified a higher rate of rare CNVs, especially in genes earlier implicated in NDDs and development of the nervous system in ASD-concordant pairs (Figure 2). The rate of putatively clinical significant CNVs in our twins concordant for NDD (13.6%) was similar to the reported rates in ASD cohorts ranging from ~10% to 28% (Ho et al., Reference Ho, Wassman, Baxter, Hensel, Martin, Prasad and Butler2016; Shen et al., Reference Shen, Dies, Holm, Bridgemohan, Sobeih, Caronna and Miller2010; Tammimies et al., Reference Tammimies, Marshall, Walker, Kaur, Thiruvahindrapuram, Lionel and Fernandez2015). Our data also suggest that there could be an increased rate of the CNVs affecting genes involved in alterations of the nervous system also in the ASD/NDD-discordant MZ pairs (Figure 2B, D) demonstrating that CNVs with incomplete penetrance could contribute to the NDD phenotypes seen in the twins. Many of the most common CNVs found in NDD cases have variable expressivity as well as incomplete penetrance (Carter & Scherer, Reference Carter and Scherer2013). More severe phenotypes connected to the recurrent genomic disorders and rare CNVs are most likely due to additional genetic variants, epigenetic modifications, environmental exposures, and their combined effects. For instance, additional rare CNVs were more common in carriers of the recurrent 16p11.2 deletions with more severe NDD phenotypes (Girirajan et al., Reference Girirajan, Rosenfeld, Cooper, Antonacci, Siswara, Itsara and Eichler2010). The benefit of MZ discordant twin pairs design is the possibility to pinpoint additional factors such as non-shared environmental factors that contribute to the etiology of NDDs alone or as modifying the effect of rare CNVs. Indeed, we have identified non-shared medical and environmental factors that are contributing to the phenotypes using this same sample of twins (Arora et al., Reference Arora, Reichenberg, Willfors, Austin, Gennings, Berggren and Bolte2017; Willfors et al., Reference Willfors, Carlsson, Anderlid, Nordgren, Kostrzewa, Berggren and Bölte2017).
Multiple studies have highlighted the need for a cross-disorder approach in genetic studies as earlier research has shown the clinical and genetic overlap between different NDDs (Lee et al., Reference Lee, Ripke, Neale, Faraone, Purcell, Perlis and Wray2013; Thapar et al., Reference Thapar, Cooper and Rutter2017). We used two different approaches, either clinical ASD diagnosis only or the umbrella category of NDD, to define the concordance and discordance of the twins for our analysis of shared CNVs. Using this approach, we demonstrate a rising degree of the rate of CNVs affecting specific genes from TD to ASD-discordant and -concordant pairs. When using the NDD concept, we did not find any differences among the NDD discordant and concordant pairs despite an elevated rate seen in comparison with the TD pairs. The lack of difference could be due to small sample size but also the effect of the milder phenotypes such as learning disabilities included in the concordant NDD group lowering the proportion of more severely affected twin pairs in the concordant category. It has been implicated that CNV burden is not increased in milder NDDs (Girirajan et al., Reference Girirajan, Brkanac, Coe, Baker, Vives, Vu and Eichler2011).
In conclusion, we add to the growing literature that postzygotic de novo CNV events in MZ twin pairs are rare and do not explain the discordance of NDD phenotypes in the studied twin pairs. We suggest that shared CNVs overlapping genes essential for brain development increase the risk for NDDs even in discordant MZ pairs. Further studies using larger twin cohorts, additional sources of DNA, and better variant detection resolution such as whole genome sequencing (Yuen et al., Reference Yuen, Merico, Bookman, Howe, Thiruvahindrapuram, Patel and Scherer2017) are needed to confirm our results and overcome the limitations of this study. Additionally, studies should aim at investigating the contribution of other risk or resilience factors contributing to the phenotypic outcomes in genetic risk backgrounds.
Declarations
Ethics Approval and Consent to Participate
This study was approved by the National Swedish and the regional ethical review board at Karolinska Institutet and by the Swedish Data Inspection Board. Written informed consent was obtained from the twins (for twins older than 15 years) and/or their parents/legal guardians. The family identifiers have been randomized for this study and do not correspond to the identifiers assigned during the recruitment in the RATSS study.
Consent for Publication
Twins (for twins older than 15 years) and/or their parents/legal guardians have given informed written consent for publication and distribution.
Availability of Data and Material
The raw datasets generated and analyzed during the current study are not publicly available because they contain private patient health information, but are available from the corresponding author (kristiina.tammimies@ki.se) upon reasonable request and subject to necessary clearances. The minimal data set needed to reproduce the results is provided in the supplement.
Competing Interests
Sven Bölte declares no direct conflict of interest related to this article. He discloses that he has in the last five years acted as an author, consultant or lecturer for Shire, Medice, Roche, Eli Lilly, Prima Psychiatry, GLGroup, System Analytic, Kompetento, Expo Medica, and Prophase. He receives royalties for textbooks and diagnostic tools from Huber/Hogrefe, Kohlhammer, and UTB. Stephen Scherer is on the Scientific Advisory Boards of Population Diagnostics, Deep Genomics, DNAStack, and has intellectual property licensed relevant to CNVs through the Hospital for Sick Children to Lineagen. The other authors declare that they have no competing interests.
Funding
Support was provided by the Innovative Medicines Initiatives Joint Undertaking (grant agreement number 115300), which comprises financial contribution from the European Union's Seventh Framework Programme (FP7/2007–2013) and in-kind contributions from companies belonging to the European Federation of Pharmaceutical Industries and Associations; the Swedish Research Council (523-2009-7054; 521-2013-2531; 350-2012-286); the Swedish Research Council, in partnership with the Swedish Research Council for Health, Working Life and Welfare, Formas and VINNOVA (cross-disciplinary research program concerning children's and young people's mental health, 259-2012-24), Stockholm County Council (20100096, 20110602, 20120067, 20140134), Stiftelsen Frimurare Barnhuset, Sunnerdahls-Handikappfond, Hjärnfonden. S.W.S. is supported by the GlaxoSmithKline-Canadian Institutes of Health (CIHR) Endowed Chair in Genome Sciences at The Hospital for Sick Children and University of Toronto. K.T is financially supported by the Swedish Foundation for Strategic Research, the Harald and Greta Jeanssons Foundations, Åke Wiberg Foundation, The Swedish Foundation for International Cooperation in Research and Higher Education, StratNeuro, The Board of Research at Karolinska Institutet and Foundations and Funds at Karolinska Institutet.
Authors’ Contributions
S.S., S.Bö., and K.T conceived and designed the study. B-M.A., C.W., S.Be., and A.N. recruited, diagnosed, and examined the twins. S.S., B.T., J.W., and K.T. processed the microarray data. S.S. and K.T. analyzed the data and drafted the manuscript. All authors contributed to acquisition of data, interpretation of results and critical discussion and approved the final version of the manuscript.
Acknowledgments
We thank all the twins and their families for participating in this study. We also thank Kerstin Andersson, Christina Coco, Martin Hammar, Johanna Ingvarsson, Anna Lange Nilsson, Gunnel Ahréns, Lynnea Myers, Ielyzaveta Rabkina, Lina Poltrago, and Anna Råde for their valuable contribution to the work presented in this study. Genotyping was performed by the SNP&SEQ Technology Platform in Uppsala (www.genotyping.se). The facility is part of the National Genomics Infrastructure (NGI) Sweden and Science for Life Laboratory. The SNP&SEQ Platform is also supported by the Swedish Research Council and the Knut and Alice Wallenberg Foundation. The computation resources were provided by SNIC through Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) (b2015257).
Supplementary material
Supplementary material is available on the Cambridge Core website.
To view supplementary material for this article, please visit https://doi.org/10.1017/thg.2017.69





