Introduction
Species distribution data may be used in assessing a species’ risk of extinction (Mace & Lande, Reference Mace and Lande1991; Rodrigues et al., Reference Rodrigues, Pilgrim, Lamoreux, Hoffmann and Brooks2006; Baillie et al., Reference Baillie, Collen, Amin, Akcakaya, Butchart and Brummitt2008; Mace et al., Reference Mace, Collar, Gaston, Hilton-Taylor, Akçakaya and Leader-Williams2008). The IUCN Red List of Threatened Species (IUCN, 2012) is a set of biodiversity data that are used to categorize species according to threats to their population, distribution and habitat (IUCN, 2001; IUCN Standards and Petitions Working Group, 2008). The Red List is used to inform policy and decision-making processes on national and international levels (Lamoreux et al., Reference Lamoreux, Akçakaya, Bennun, Collar, Boitani and Brackett2003; Mace & Baillie, Reference Mace and Baillie2007; Hoffmann et al., Reference Hoffmann, Brooks, da Fonseca, Gascon, Hawkins and James2008; Nicholson et al., Reference Nicholson, Collen, Barausse, Blanchard, Costelloe and Sullivan2012) and in biogeographical and biodiversity priority-setting analyses (e.g. Stuart et al., Reference Stuart, Chanson, Cox, Young, Rodrigues, Fischman and Waller2004; Grenyer et al., Reference Grenyer, Orme, Jackson, Thomas, Davies and Davies2006; Isaac et al., Reference Isaac, Turvey, Collen, Waterman and Baillie2007; Schipper et al., Reference Schipper, Chanson, Chiozza, Cox, Hoffmann and Katariya2008).
Of the estimated 5 ± 3 million species, of which 1.5 million have been described scientifically (Costello et al., Reference Costello, May and Stork2013), 63,837 (primarily vertebrates) are included on the 2012 IUCN Red List. Of the reptile species described, 38% (3,363) have been assessed and 802 species are categorized as threatened (IUCN, 2012). Approximately half of all chameleon species (the majority from Madagascar) have been assessed using the Red List criteria.
Red List assessments are typically completed at a workshop, where experts synthesize published and unpublished data within a formalized Species Information System. This approach is designed to overcome limitations in the availability of data, allows screening of existing data and compiles accumulated field experience that is not available in any published format (Huang et al., Reference Huang, Hawkins, Lei, Miller, Favret, Zhang and Qiao2012).
According to estimates there may be 2–3 billion specimens in natural history collections (Duckworth et al., Reference Duckworth, Genoways and Rose1993; Ariño, Reference Ariño2010; Scoble, Reference Scoble2010), and in the past using these museum data for biodiversity research was challenging because information had to be compiled manually from the collections. However, many museums have digitized the information contained in their specimen collections and made it available through the Global Biodiversity Information Facility (GBIF, 2014), an electronic data portal that provides a single point of access to databases from hundreds of museums and private collections. Although a large amount of data is available through GBIF it is still only an estimated 3.2% of the primary biodiversity data records in collections. The reasons for this include a lack of funds for digitization, and political and social boundaries to data sharing. Consequently the records available through GBIF tend to be from larger institutions, mostly in the developed world.
The museum specimen data in GBIF have good temporal and geographical coverage (Boakes et al., Reference Boakes, McGowan, Fuller, Chang-Qing, Clark, O'Connor and Mace2010), and questionable records can be checked and details verified against the actual specimens. These data therefore have potential to support both biogeographical analyses and conservation decision-making but problems may arise as a result of inaccurate identification of specimens, outdated taxonomy, incorrect localities and poor transcribing of data from specimen labels into computerized systems (Ponder et al., Reference Ponder, Carter, Flemons and Chapman2001; Ariño, Reference Ariño2010; Newbold, Reference Newbold2010).
GBIF data are increasingly being used in studies of species richness and endemism (Foley et al., Reference Foley, Weitzman, Miller, Faran, Rueda and Wilkerson2008; Désamoré et al., Reference Désamoré, Laenen, González-Mancebo, Jaén Molina, Bystriakova and Martinez-Klimova2012; Ramirez-Villegas et al., Reference Ramirez-Villegas, Jarvis and Touval2012) and as presence data in species distribution modelling (Costa et al., Reference Costa, Nogueira, Machado and Colli2010; Edvardsen et al., Reference Edvardsen, Bakkestuen and Halvorsen2011; Vidal-Garcia & Serio-Silva, Reference Vidal-García and Serio-Silva2011; Willis et al., Reference Willis, Jeffers, Tovar, Long, Caithness and Smit2012) but are not yet being used in Red List assessment workshops for animals, although herbarium records and databases such as Tropicos (2014) are used for Red List assessments of plants (Willis et al., Reference Willis, Moat and Paton2003; Raimondo, Reference Raimondo2009; Rivers et al., Reference Rivers, Taylor, Brummitt, Meagher, Roberts and Nic Lughadha2011).
Here we compare basic data quality attributes for 35 chameleon species endemic to Kenya and Tanzania (Fig. 1), using a GBIF dataset and expert data compiled from museum records, field research records and literature. Both datasets are then used to estimate the Extent of Occurrence (EOO, the area that encompasses all known occurrences) and Area of Occupancy (AOO, the area within the EOO that contains suitable habitat), which are the standard measures of distribution used in Red List assessments. The estimated EOO and AOO from each dataset are then used as the sole inputs to a hypothetical IUCN Red List assessment process, to compare similarities and differences in Red List categorization using each dataset. The full Red List assessment process would use a more complete set of data than range size but our aim was to illustrate differences that can arise when using different data sources.
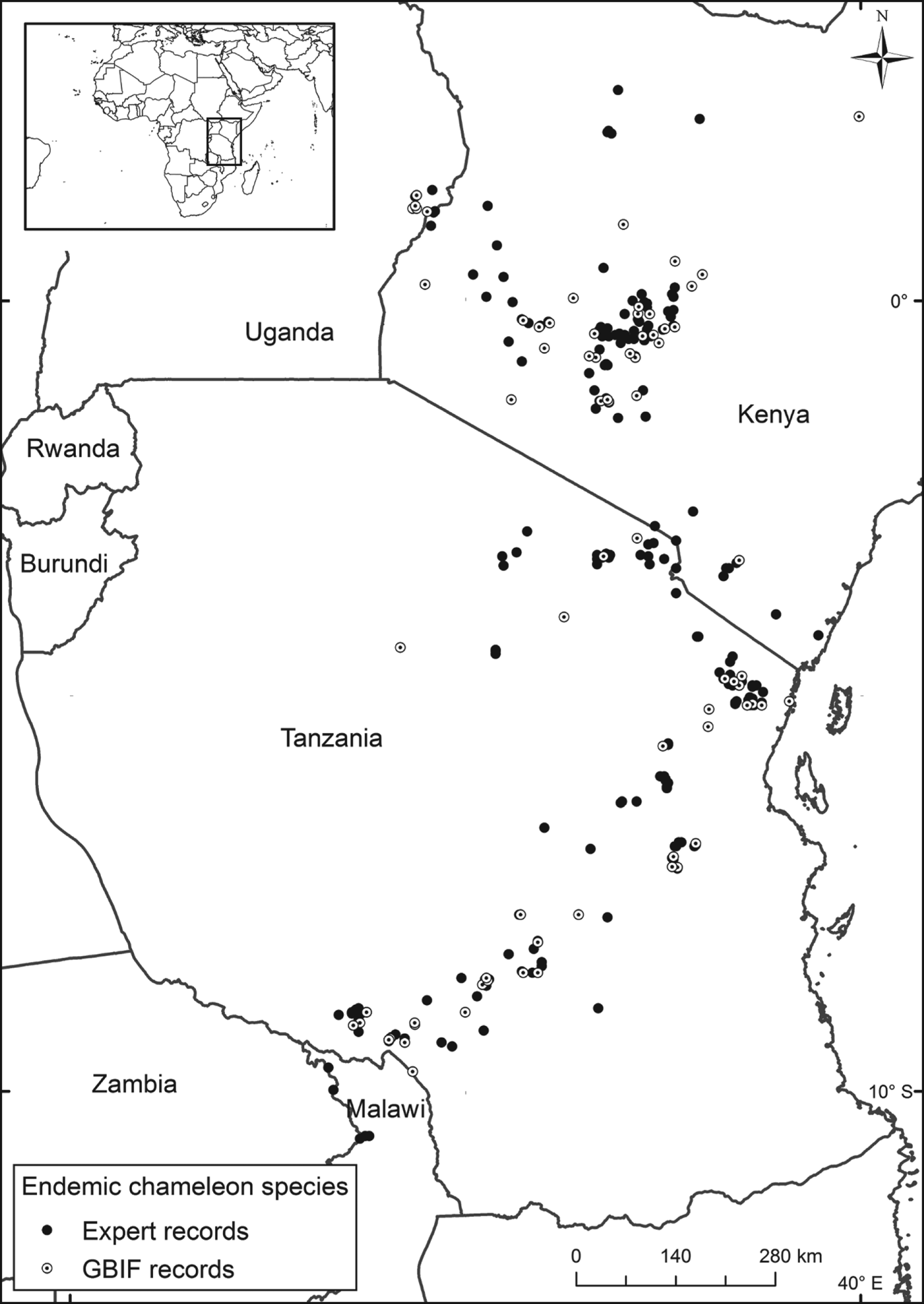
Fig. 1 Location of records of endemic East African chameleon species according to the expert (254 records) and GBIF (172 records) datasets. The rectangle on the inset indicates the location of the main map in Africa.
Methods
We compiled a list of endemic chameleon species in Kenya and Tanzania, which was cross-checked by members of the IUCN Chameleon Specialist Group. Species assignments followed the most recent accepted taxonomy (Tilbury, Reference Tilbury2010).
From this list a sample of 35 species was selected based on data availability. We created two datasets for the sample, one with expert data compiled and vetted by chameleon expert Colin Tilbury and the other with raw data obtained through GBIF. The expert dataset comprised 263 records assembled for the production of an atlas of African chameleons (Tilbury, Reference Tilbury2010), which were sourced from museums, peer-reviewed literature and unpublished field observations. The quality of locality information was checked by the expert and taxonomic data were updated. The second dataset comprised 2,304 museum records sourced from the GBIF data portal, using a query for records of chameleons in Kenya and Tanzania, which were then filtered to include only endemic species. These records were from six museums: Harvard Museum of Comparative Zoology, USA; Smithsonian Institution, USA; Ditsong Museum, South Africa; Bishop Museum, USA; Los Angeles County Museum of Natural History, USA; and California Academy of Sciences, USA. The expert dataset included data from 11 museums. The only museum included in both datasets was Ditsong Museum (Table 1).
Table 1 Museums from which locality records for East African chameleons were obtained for the expert and GBIF datasets.

Records from both datasets were cleaned according to criteria described in Chapman (Reference Chapman2005), which included the removal of all records without geographical coordinates, all duplicate records and all specimens not identified to the species level. Scientific names were standardized against the currently accepted taxonomy (Table 2) and coordinates were checked in ArcGIS v. 10.1 (ESRI, Redlands, USA), with the removal of obvious outlier data points. Following this cleaning process 254 expert records (93.3% of the original expert dataset) and 172 GBIF records (7.5% of the original GBIF dataset) were retained for further analysis.
Table 2 Data quality and data cleaning requirements for the expert and GBIF databases, with numbers of records of raw data, cleaned data, requiring taxonomic update, for Chamaeleo species, with no geographical coordinates, and with no locality listed. Total cleaned data were the records used in the analysis.

* Many species in this genus have been reassigned to Kinyongia and Trioceros, but these changes are not reflected in the GBIF dataset.
Once cleaned, distribution records for every species in each dataset were imported into ArcGIS, which was used to estimate the EOO and AOO. The EOO was estimated by creating convex hull polygons, using the point data records, to represent the potential habitat range for each species. The AOO was estimated using the guidelines provided by the IUCN Standards and Petitions Working Group (2008), using Jenness repeating shapes (Jenness, Reference Jenness2012) to create a fishnet grid of 4-km2 hexagons. This is the scale recommended by the IUCN. The majority of the records did not have a precision estimate nor did they indicate the datum used when recording the locality coordinates. For mapping purposes it was assumed that WGS 84 was used as it is the default datum used in global positioning systems.
Criterion B of the Red List Categories and Criteria version 3.1 (geographical range in the form of EOO or AOO; IUCN, 2001) was applied to the calculated EOO/AOO for each species and used to categorize species as Critically Endangered, Endangered or Vulnerable. If there were not at least three locality records, which are required to estimate EOO, the taxon was categorized as Data Deficient. No other criteria (e.g. fragmentation, number of locations, decline in habitat) were considered.
Results
Almost all of the records (99.9%) in the GBIF dataset had taxonomy that did not match the species in our checklist, principally because the records accessed through GBIF were not updated to generic-level reassignments made within Chamaeleonidae during 2006–2013. In the GBIF dataset 478 of the records (21%) had no geographical coordinates, whereas only seven records (3%) in the expert dataset lacked coordinates. In addition, 833 records (36%) in the GBIF dataset had no locality name, whereas all records in the expert dataset had a locality name. Records without locality but with coordinates were used in the analysis but it was not possible to verify the accuracy of these records.
Some records for Fischer's chameleon Kinyongia fischeri, Von Hohnel's chameleon Trioceros hoehnelii and Jackson's chameleon Trioceros jacksonii in the GBIF dataset were regarded as outliers because they were far outside known distributions. For K. fischeri the suspected outlier was 221 km from the nearest record in the expert dataset and 160 km from the nearest record in the GBIF dataset. There was 48 km between the nearest points in the GBIF and expert datasets (Fig. 2). In the GBIF dataset one record for T. hoehnelii was 218 km from other T. hoehnelii records and one record for T. jacksonii was 1,097 km from other T. jacksonii records. These two records probably resulted from misidentification.

Fig. 2 Extent of occurrence (EOO) of Kinyongia fischeri in Tanzania, calculated using expert and GBIF datasets.
In the expert dataset there were sufficient distribution records to estimate the EOO for 26 species and the AOO for 34 species (Table 3). The EOO could not be estimated for Kinyongia magomberae, K. uluguruensis, K. vanheygeni, K. vosseleri, Rhampholeon acuminatus, R. beraduccii, Trioceros marsabitensis, T. narraioca and T. ntunte because there were only one or two records for these species. In the GBIF dataset there were sufficient data to estimate the EOO for 10 species and the AOO for 19 species.
Table 3 Extent of occurrence (EOO) calculated using expert and GBIF datasets, area of overlap between the databases, and percentage overlap for each database, for 10 species of East African chameleon.

The EOO for each species had varying degrees of overlap between the two datasets. For 10 of the 35 species there were overlaps in the ranges calculated using the GBIF and expert datasets (Table 3), with T. deremensis, T. tempeli and T. hoehnelii overlapping by > 50%. For the remaining species there were large differences in the ranges calculated using the expert and GBIF datasets, which resulted in overlaps of < 50% and in some cases 0%. When outliers were removed there were no range overlaps between the datasets for K. fischeri (Fig. 2) and K. excubitor (Fig. 3). The range for T. goetzei calculated using data from GBIF was located 100% within the range calculated using the expert dataset. The range for T. jacksonii calculated using the expert dataset was located 100% within the range calculated using the GBIF dataset.

Fig. 3 EOO of Kinyongia excubitor in Kenya, calculated using expert and GBIF datasets.
Using the expert dataset 34 of the species were assigned a threat category, with only T. ntunte categorized as Data Deficient, whereas using the GBIF dataset 16 species (46%) were categorized as Data Deficient. None of the species were categorized as Vulnerable, Near Threatened or Least Concern as all ranges were smaller than the minimum areas required for these categories.
The differences in geographical data for species in the two datasets resulted in some disparity in the Red List threat category assignments (Table 4). Only 10 (29%) species had sufficiently similar EOO/AOO to be assigned the same Red List category for both datasets. For eight species the differences in the EOO/AOO between the datasets placed them in different threat categories under the Red List system, assuming additional criteria were met. Using the expert dataset these species would be categorized as Endangered, whereas using the GBIF dataset they would be categorized as Critically Endangered.
Table 4 Potential IUCN Red List categories that might be assigned to East African chameleon species, based on assessments using expert and GBIF datasets.

* CR, Critically Endangered; EN, Endangered; VU, Vulnerable; DD, Data Deficient
Discussion
These results show that distribution data for endemic East African chameleons vary substantially between expert and GBIF data sources, and this disparity would result in species being assigned to different IUCN Red List threat categories based on Criterion B. Less than 30% of the comparisons produced congruent results, with highly variable amounts of overlap between the ranges estimated using our two datasets.
Our results show that the quality of the raw GBIF dataset for East African chameleons is insufficient for direct use in assessments of IUCN threat status. There were two principal problems with the GBIF data. Firstly, most of the records obtained through GBIF used outdated taxonomy and did not reflect the numerous reassignments that have been made at the genus level. Secondly, 833 (36%) of the GBIF records had no locality listed. Without expert review it is impossible to know if a locality record is correct.
Before being suitable for use in a Red List assessment, GBIF data will require basic geographical analysis such as checking for incorrect locality records and taxonomic updates. In comparison, our expert dataset was essentially ready for use in an IUCN Red-listing task. The perceived added value from including a large number of specimen records in the GBIF database was marginal because of the poor quality of the specimen records. Two other studies evaluating the fitness-for-use of GBIF data confirm that these problems also exist for other taxa, on a regional and global scale (Gaiji et al., Reference Gaiji, Chavan, Ariño, Otegui, Hobern, Sood and Robles2013; Otegui et al., Reference Otegui, Ariño, Encinas and Pando2013).
The taxonomic shortcomings that we identified in the GBIF data are being addressed; for example taxonstand draws on a standardized database of plant names, and updates taxonomy automatically within the R environment (Cayuela et al., Reference Cayuela, Granzow-de la Cerda, Albuquerque and Golicher2012). Until additional resources are available to automate this process locality records should be vetted against current taxonomy and, if possible, verified by taxonomic experts. Following agreed standards for the georeferencing of localities data from museum specimen tags (Chapman & Wieczorek, Reference Chapman and Wieczorek2006) improves the usability of data but is expensive and time consuming.
Some of the differences between the datasets could also be attributed to biases, which are common in biodiversity research. For example, species records can be biased by expert knowledge (Ahrends et al., Reference Ahrends, Rahbek, Bulling, Burgess, Platts and Lovett2011b), accessibility (Freitag et al., Reference Freitag, Hobson, Biggs and van Jaarsveld1998; Kadmon et al., Reference Kadmon, Farber and Danin2004) and the popularity of a location for researchers (Reddy & Dávalos, Reference Reddy and Dávalos2003; Ahrends et al., Reference Ahrends, Burgess, Gereau, Marchant, Bulling and Lovett2011a).
An important final question is whether either dataset provides sufficient coverage for accurate Red List assessments. In our view there were sufficient records to estimate the EOO/AOO for most species. However, differences between the two datasets could result in the assignment of species to different IUCN categories and this could have significant consequences as conservation decisions are often based on the IUCN Red List.
Despite the challenges faced when using GBIF data for Red List assessment, this public database is useful for filling knowledge gaps and facilitating the development of provisional maps of distribution, which can then be used in preliminary analyses such as those presented here, checked by experts, and used to identify potentially problematic issues in the estimation of EOO and AOO. Furthermore, large compilations of museum specimen data may be useful for macroecological analyses (Willis et al., Reference Willis, Jeffers, Tovar, Long, Caithness and Smit2012).
This exploratory exercise yielded a number of conclusions. Firstly, for East African chameleon species GBIF data are insufficient for Red List assessments because of problems with taxonomy and data outliers. Secondly, once GBIF data are vetted against current taxonomy and species localities are verified they are adequate to estimate the EOO for most species. Thirdly, gaps in GBIF data coverage may result in a species being assigned a higher category of threat in IUCN Red List assessments. We conclude that there is no substitute for the taxonomic expert in data compilation and analysis processes such as Red List assessments.
Acknowledgements
We thank Colin Tilbury for the use of his data in this study, Samy Gaiji and Vishwas Chavan from the Global Biodiversity Information Facility and Richard Jenkins from the IUCN Global Species Programme for support and comments, and the Chameleon Specialist Group for their assistance with verifying our species checklist.
Biographical sketches
Angelique Hjarding is interested in biogeography, landscape ecology, urbanization and farmland preservation. Krystal A. Tolley specializes in chameleon biogeography and systematics. Neil D. Burgess is a conservation scientist specializing in African biological conservation issues, particularly in Eastern Africa.









