Article contents
Nanostructured substrates for multi-cue investigations of single cells
Published online by Cambridge University Press: 30 January 2018
Abstract
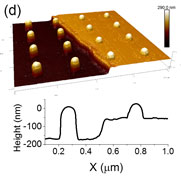
Cellular adhesion depends on the integration of numerous signaling inputs generated by the chemical and physical properties of the substrate. The complex coupling among inputs makes it challenging experimentally to deconvolve their individual contributions to the adhesion process. To address this roadblock, we have employed a combination of electron beam and optical lithographic techniques to fabricate substrates with independently tunable topographical and chemical signaling cues. Arrays of gold nanostructures were patterned atop quartz substrates, half of which were etched into gold-capped nanopillars. Individual A549 cells exposed simultaneously to Arg-Gly-Asp-functionalized etched and non-etched arrays exhibited strongly preferential adherence to the nanopillars.
- Type
- Research Letters
- Information
- Copyright
- Copyright © Materials Research Society 2018
References
- 5
- Cited by


