Introduction
The complicated taxonomic history of the species of the genus Serrasentis Van Cleave 1923 (Echinorhynchida: Isthmosacanthidae) has been outlined and analysed in different studies (i.e., Gupta and Jain Reference Gupta and Jain1985; Bhattacharya Reference Bhattacharya2007; Amin Reference Amin2013; Barton et al. Reference Barton, Smales and Morgan2018; Amin and Heckmann Reference Amin and Heckmann2021), showing that the number of valid species has been a source of debate. According to Barton et al. (Reference Barton, Smales and Morgan2018), who thoroughly revised a list of fourteen species by Amin (Reference Amin2013), of the fourteen species, three (Serrasentis chauhani Datta, 1954; Serrasentis fotedari Gupta & Fatma, 1980; and Serrasentis golvani Gupta & Kumar, 1987) are junior synonyms of Serrasentis sagittifer (Linton, 1889) Van Cleave, 1932, and three (Serrasentis engraulisi Gupta & Gupta, 1980; Serrasentis mujibi Bilqees, 1972; and Serrasentis psenesi Gupta & Gupta, 1980) of Serrasentis nadakali George & Nadakal, 1978, based on different studies (Gupta and Jain Reference Gupta and Jain1985; Bhattacharya Reference Bhattacharya2007; Barton et al. Reference Barton, Smales and Morgan2018). Additionally, due to inadequate species descriptions based on only one larval stage and/or an insufficient sample size, Barton et al. (Reference Barton, Smales and Morgan2018) designated five as species inquirenda, or uncertain (Serrasentis longiformis Bilqees, 1974; Serrasentis niger Khatoon & Bilqees, 2007; Serrasentis sidaroszakaio Tadros, Iskandar & Wassef, 1979; Serrasentis sauridae Surekha & Vijayalakshmi, 2006; Serrasentis sciaenus Bilqees, 1972), four of which Amin (Reference Amin2013) had considered valid. Barton et al. (Reference Barton, Smales and Morgan2018) also recognized one valid species that was not included in Amin’s (Reference Amin2013) list, namely Serrasentis indicus Singh, Agarwal & Lakshmi, Reference Singh, Agrawal and Anuradha1998. Thus, at the time of Barton et al.’s (Reference Barton, Smales and Morgan2018) publication, the genus contained five valid species. An additional species was described by Gupta (Reference Gupta2021), whereby the genus Serrasentis now contains the following six valid species (Barton et al. Reference Barton, Smales and Morgan2018; WoRMS 2023): Serrasentis indica Singh, Agarwal & Lakshmi, Reference Singh, Agrawal and Anuradha1998; Serrasentis lamelliger (Diesing, 1854) Meyer, 1932; Serrasentis manazo Bilqees & Khan, 2005; Serrasentis nadakali George & Nadakal, 1978; Serrasentis sagittifer (Linton, 1889) Van Cleave, 1932; and Serrasentis synagrisi Gupta, Reference Gupta2021.
Serrasentis sagittifer, the type species, is the only species reported from this genus in the Americas (Barton et al. Reference Barton, Smales and Morgan2018; Amin and Heckmann Reference Amin and Heckmann2021 and citations therein). Cobia, Rachycentron canadum (Linnaeus, 1766) (Rachycentridae) is considered the primary marine definitive host of S. sagittifer (Barton et al. Reference Barton, Smales and Morgan2018; Amin and Heckmann Reference Amin and Heckmann2021). Based on the worldwide distribution and natural history of cobia, Amin and Heckmann (Reference Amin and Heckmann2021) have explained the morphological variability of S. sagittifer and the distribution of several paratenic hosts of marine waters of the Arabian Gulf, Australia, and the Americas.
Several taxa of the genus Serrasentis were reported as parasites of 10 marine teleost species included in six families in Mexican waters from the Gulf of Mexico (GoM) (see Table S1 for more details). For example, adult specimens of S. sagittifer have been reported from “cod” (Atlantic hake) and Umbrina coroides Cuvier, 1830 (Salgado-Maldonado Reference Salgado-Maldonado1978; Montoya-Mendoza et al. Reference Montoya-Mendoza, Salgado-Maldonado, Álvarez-Noguera, Lugo-Vázquez and Lango-Reynoso2019); juvenile specimens of S. sagittifer have been reported from Lutjanus campechanus (Poey, 1860) and Scorpaena mystes Jordan & Starks, 1895 (Montoya-Mendoza et al. Reference Montoya-Mendoza, Jiménez-Badillo, Salgado-Maldonado and Mendoza-Franco2014, Reference Montoya-Mendoza, Badillo-López, Amaro-Espejo, Castañeda-Chávez, Lango-Reynoso and Herrera-Martínez2018); and specimens of S. sagittifer with unspecified life-cycle stage have been reported from Bagre marinus (Mitchill, 1815) and Micropogonias undulatus (Linnaeus, 1766) from the coasts of Veracruz, Mexico (Chávez-Lopez et al. Reference Chávez-López, Montoya-Mendoza, Franco-López, Barrera-Escorcia and Morán-Silva1996; Salgado-Maldonado and Amin Reference Salgado-Maldonado, Amin, Felder and Camp2009; García-Prieto et al. Reference García-Prieto, García-Varela, Mendoza-Garfias and Pérez-Ponce de León2010). Adult specimens of S. sagittifer have been reported from Haemulon aurolineatum Cuvier, 1830 from the Yucatán Continental Shelf, Mexico (García-Teh Reference García-Teh2020). Adult Serrasentis sp., on the other hand, have been reported from Lutjanus griseus (Linnaeus, 1758) from Chelem lagoon and Progreso port (coastal area), Yucatán, Mexico (Argáez-García et al. Reference Argáez-García, Guillén-Hernández and Aguirre-Macedo2010), and unspecified life-cycle stage Serrasentis sp. have been reported from Micropogonias undulatus (Linnaeus, 1766) from Laguna Madre, Tamaulipas, Mexico (Iruegas-Buentello Reference Iruegas-Buentello1999). Particularly, recent studies of helminth communities from intestines of three flatfish species (Paralichthyidae) (i.e., Cyclopsetta chittendeni Bean, 1895; Syacium gunteri Ginsburg, 1933; and Syacium papillosum (Linnaeus, 1758)) from oceanic sites in the southern Gulf of Mexico (s-GoM) have reported S. sagittifer as the most prevalent adult acanthocephalan parasite species (Vidal-Martínez et al. Reference Vidal-Martínez, Centeno-Chalé, Torres-Irineo, Sánchez-Ávila, Gold-Bouchot and Aguirre-Macedo2014, Reference Vidal-Martínez, Velázquez-Abunader, Centeno-Chalé, May-Tec, Soler-Jiménez, Pech, Mariño-Tapia, Enriquez, Zapata-Pérez, Herrera-Silveira, Hernández-Mena, Herzka, Ordoñez-López and Leopoldina Aguirre-Macedo2019; Centeno-Chalé et al. Reference Centeno-Chalé, Aguirre-Macedo, Gold-Bouchot and Vidal-Martínez2015).
The universal molecular marker ‘DNA barcoding’ (a fragment of the mitochondrial cytochrome C oxidase subunit I gene [=mtDNA COI region barcoding, henceforth referred to as barcoding] [Hebert et al. Reference Hebert, Cywinska, Ball and de Waard2003]), together with morphological evidence, has been used for the identification and discovery of new species of acanthocephalans from marine teleosts (e.g., Braicovich et al. Reference Braicovich, Lanfranchi, Farber, Marvaldi, Luque and Timi2014; Lisitsyna et al. Reference Lisitsyna, Scholz and Kuchta2015, Reference Lisitsyna, Kudlai, Cribb and Smit2019a, Reference Lisitsyna, Kudlai, Spraker, Tkach, Smales and Kuzminab; Amin et al. Reference Amin, Heckmann, Dallarés, Constenla and Ha2019; Huston and Smales Reference Huston and Smales2021; Kaur et al. Reference Kaur, Shamal, Chandran, Krupesha Sharma and Sanil2021). However, barcoding data from Isthmosacanthidae acanthocephalans are scarce (Huston et al. Reference Huston, Cribb and Smales2020a, Reference Huston, Cribb and Smalesb; Huston and Smales Reference Huston and Smales2020, Reference Huston and Smales2021).
As a part of an ongoing study to document the baseline biodiversity of the GoM (Consorcio de Investigación del Golfo de Mexico [CIGoM]; www.cigom.org, last accessed 1 December 2023), with special emphasis on parasite diversity, several acanthocephalans belonging to the genus Serrasentis were collected from the intestines of four flatfish species (Paralichthyidae) namely Ancyclopsetta quadrocellata Gill, 1864; C. chittendeni; S. gunteri; and S. papillosum from 10 offshore sites in the GoM. An examination of these specimens based on an integrative taxonomic approach revealed that they belong to a new species of the genus Serrasentis, which is described here. In this context, the aims of this study were to 1) provide a detailed morphological description of the specimens of Serrasentis and offer new morphometric and molecular data (i.e., DNA sequences) to facilitate future inter and intra-specific systematic comparisons in Serrasentis; 2) provide images of scanning electron microscopy (SEM) for the specimens of Serrasentis collected in this study; 3) use genetic (barcoding sequence) data to determine sister-group relations of Serrasentis used in this study within a phylogenetic framework of the Isthmosacanthidae; and 4) explore the intraspecific genetic variation of Serrasentis from the GoM.
Materials and methods
Collection of flatfishes, tomtate grunt, and acanthocephalans
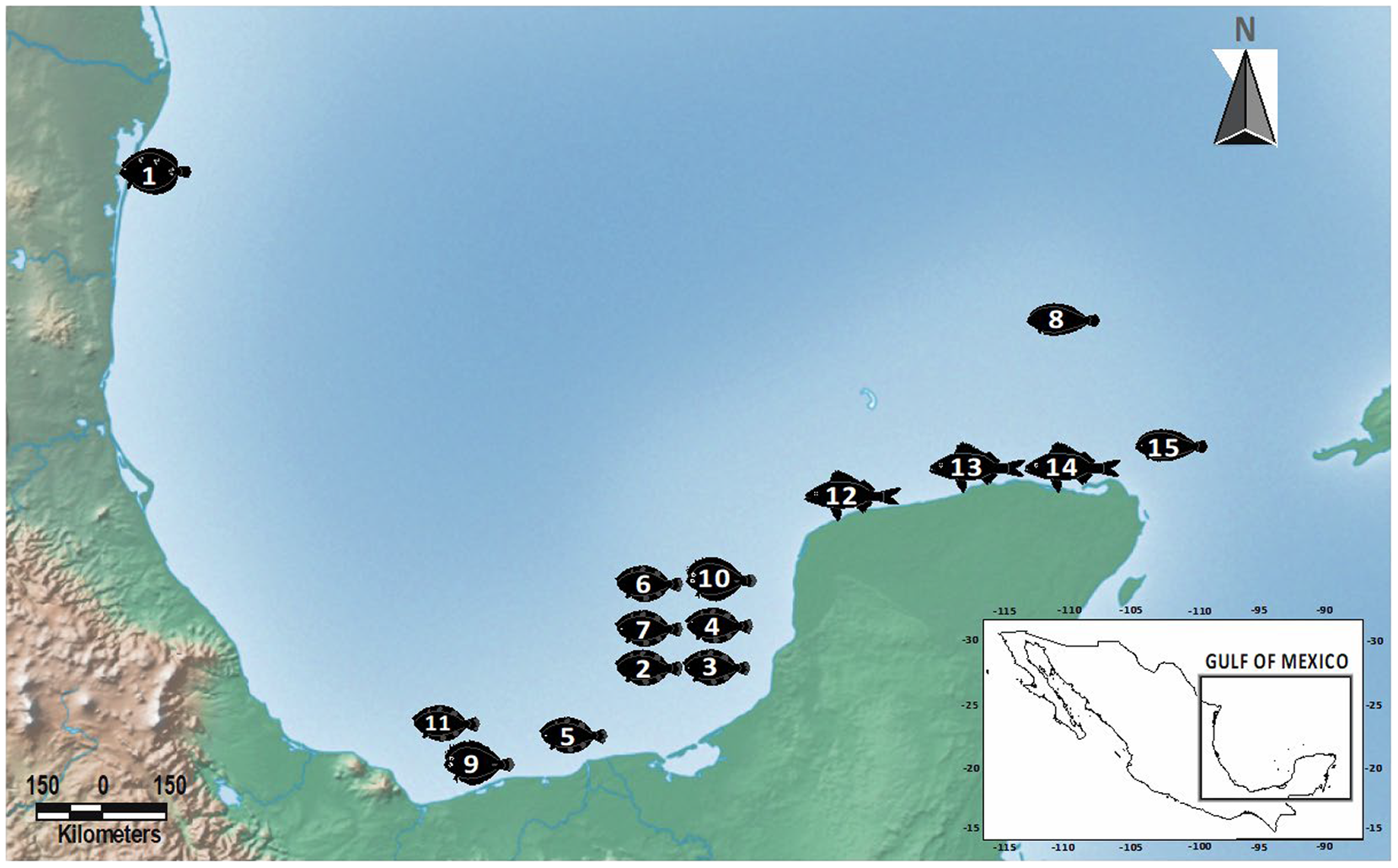
Adult acanthocephalans of the genus Serrasentis were collected from the intestines of four flatfish species (Paralichthyidae), namely A. quadrocellata (depth 51 m); C. chittendeni (depth range 29–68 m); S. gunteri (depth range 27–36 m); and S. papillosum (Linnaeus, 1758) (depth 100 m). Host species were collected from one, six, two, and one oceanic sampling sites, respectively. Additionally, adult specimens of Gorgorhynchus lepidus Van Cleave, 1940 (Rhadinorhynchidae: Gorgorhynchinae) were collected from the intestine of C. chittendeni (depth 46 m), S. papillosum (depth 79 m), and one species of Haemulidae, the tomtate grunt Haemulom aurolineatum Cuvier, 1830 (depth range 14–22 m) from one, one and three sampling sites, respectively (Table 1, Figure 1).
Table 1. Collection data for acanthocephalan species sequenced in this study. IN, individual host number; #OSS, oceanic sampling site number; SD, sea depth (meters); GenBank, GenBank accession number of barcoding sequences generate in this study (– = not sequenced); CHCM, voucher numbers of individuals deposited at Colección Helmintológica del CINVESTAV; P, Paralichthydae; H, Haemulidae; Hg, hologenophore; Pg, paragenophore (terminology follows Pleijel et al. Reference Pleijel, Jondelius, Norlinder, Nygren, Oxelman, Schander, Sundberg and Thollesson2008)

The sampling was carried out at selected sites within a polygon with a total area of 341,824.94 km2 in the GoM. Samples were obtained from March 2015 to June 2021. Oceanographic sampling procedures for the collection of fishes have been described elsewhere (i.e., Vidal-Martínez et al. Reference Vidal-Martínez, Centeno-Chalé, Torres-Irineo, Sánchez-Ávila, Gold-Bouchot and Aguirre-Macedo2014, Reference Vidal-Martínez, Velázquez-Abunader, Centeno-Chalé, May-Tec, Soler-Jiménez, Pech, Mariño-Tapia, Enriquez, Zapata-Pérez, Herrera-Silveira, Hernández-Mena, Herzka, Ordoñez-López and Leopoldina Aguirre-Macedo2019; García-Teh et al. Reference García-Teh, Vidal‑Martínez, Mariño‑Tapia, Árcega‑Cabrera, Ordoñez‑López and Aguirre‑Macedo2022). Host dissection procedures followed Vidal-Martínez et al. (Reference Vidal-Martínez, Centeno-Chalé, Torres-Irineo, Sánchez-Ávila, Gold-Bouchot and Aguirre-Macedo2014, Reference Vidal-Martínez, Velázquez-Abunader, Centeno-Chalé, May-Tec, Soler-Jiménez, Pech, Mariño-Tapia, Enriquez, Zapata-Pérez, Herrera-Silveira, Hernández-Mena, Herzka, Ordoñez-López and Leopoldina Aguirre-Macedo2019) and García-Teh et al. (Reference García-Teh, Vidal‑Martínez, Mariño‑Tapia, Árcega‑Cabrera, Ordoñez‑López and Aguirre‑Macedo2022). Acanthocephalan specimens were found attached to the intestine of the fish with the proboscis invaginated. Acanthocephalans collected were first maintained at 4°C for 12 h in distilled water to produce proboscis evagination and subsequently handled manually with fine brushes, to be fixed in 100% ethanol for morphological and/or molecular analyses. Acanthocephalans (with the invaginate proboscis) were stored directly in 96% ethanol and used for molecular analyses.
The flatfishes and tomtate grunt were collected by professional fishermen using a commercial fishing permit issued by the Secretaría de Ganadería, Desarrollo Rural, Pesca y Alimentación (number 01067, and available upon request) and by the Comisión Nacional de Acuacultura y Pesca (PPF/DGOPA-070/16). The fishing activities did not involve endangered or protected species according to Mexican regulations (NOM-059-SEMARNAT-2001).
Morphological data and morphometric analyses
Acanthocephalan specimens were stained with Meyer´s carmine, dehydrated with ethanol graduated (80%, 90%, 96%, and 100%), rinsed in different concentrations of clove oil (10%, 50%, 96%, and 100%), and mounted on permanent slides using Canada balsam (Vidal-Martínez et al. Reference Vidal-Martínez, Aguirre-Macedo, Scholz, González-Solís and Mendoza-Franco2001). Serrasentis specimens were observed and measured with an Olympus BX50 (Olympus, Tokyo, Japan) optical microscope with DIC Nomarski phase contrast, photographed on a digital camera with Evolution MP color (Media cybernetics, Rockville, Maryland USA), using Qcapture 2.98.2 software (Quantitative Imaging Corporation, Surrey, BC Canada, 2009). Serrasentis specimens’ drawings were produced with the aid of a drawing tube attached to an Olympus BX50 microscope and digitized in Adobe Illustrator 2023 (Adobe Inc, Portland, OR, USA). Morphological measurements are presented in micrometers (μm) as ranges followed by the means in parentheses. For SEM, Serrasentis specimens were fixed in glutaraldehyde 3% (24 h), dehydrated in different ethanol concentrations (30%, 50%, 70%, 90%, and 100%), dried at critical point with CO2, and covered with a layer of palladium-gold (Au-Pb). Finally, they were observed and photographed on a Philips XLE30 ESEM microscope (FEI Company, Hillsboro, Oregon, USA). Because the initial handling of the specimens was not ideal for SEM photography, and as a result the specimens in the SEM images were not in perfect shape, the observations of key structures were based on the specimens mounted in Canada balsam. Serrasentis specimens were identified following/contrasting the taxonomic criteria of Golvan (Reference Golvan1969), Salgado-Maldonado (Reference Salgado-Maldonado1978), Naidu (Reference Naidu2012), Amin (Reference Amin2013), Bilqees and Khan (Reference Bilqees and Khan2015), Barton et al. (Reference Barton, Smales and Morgan2018), Fonseca et al. (Reference Fonseca, Knoff, Felizardo, Lopes Torres, Nogueira Di Azevedo, Gomes, de São Clemente and Iñiguez2019), Gupta (Reference Gupta2021), and Amin and Heckmann (Reference Amin and Heckmann2021). Several voucher specimens were compared with the newly collected specimens, i.e., ‘Serrasentis sagittifer’ (Colección Helmintológica del CINVESTAV-Unidad Mérida [CHCM], Departamento de Recursos del Mar, Centro de Investigación y de Estudios Avanzados del Instituto Politécnico Nacional [No. CHCM 606]; Colección Nacional de Helmintos [CNHE], Instituto de Biología, Universidad Nacional Autónoma de Mexico [UNAM] [No. CNHE 9352]; ‘Serrasentis sagittifer’ is mentioned in quotes due to the misidentification of these specimens from Vidal-Martínez et al. (Reference Vidal-Martínez, Centeno-Chalé, Torres-Irineo, Sánchez-Ávila, Gold-Bouchot and Aguirre-Macedo2014, Reference Vidal-Martínez, Velázquez-Abunader, Centeno-Chalé, May-Tec, Soler-Jiménez, Pech, Mariño-Tapia, Enriquez, Zapata-Pérez, Herrera-Silveira, Hernández-Mena, Herzka, Ordoñez-López and Leopoldina Aguirre-Macedo2019) and Gorgorhynchus lepidus Van Cleave, 1940 (No. CHCM 607).
Several acanthocephalan specimens chosen for molecular and morphological analysis were designated as vouchers according to Pleijel et al. (Reference Pleijel, Jondelius, Norlinder, Nygren, Oxelman, Schander, Sundberg and Thollesson2008) as follows. For Serrasentis, paragenophores (different individuals obtained from the same host and/or location and/or sampling event) of specimens used for molecular analyses were processed for morphological analysis and used as voucher specimens. For G. lepidus, the body (without proboscis) of each selected specimen was used for DNA extraction, and the remaining part of the individual (hologenophore) was used as a voucher specimen (i.e., evaginated proboscis). The voucher specimens were deposited in the CHCM (Table 1).
DNA extraction, PCR, and sequencing
Deoxyribonucleic acid (DNA) was extracted from 25 adult acanthocephalans (for details see Table 1), using the DNeasy blood and tissue extraction kit (Qiagen, Valencia, CA, USA) following the manufacturer’s instructions. The barcoding region was amplified by Polymerase Chain Reaction (PCR) (Saiki et al. Reference Saiki, Gelfand, Stoffel, Scharf, Higuchi, Horn, Mullis and Erlich1988), using the primers #507 forward (5´ - AGT TCT AAT CAT AA(R) GAT AT(Y) GG - 3´) (Nadler et al. Reference Nadler, Bolotin and Stock2006) and HCO2198 reverse (5´ - TAA ACT TCA GGG TGA CCA AAA AAT CA - 3´) (Folmer et al. Reference Folmer, Black, Hoeh, Lutz and Vrijenhoek1994). The reactions were prepared using the Green GoTaq Master Mix (Promega Inc., Madison, WI, USA). This procedure was carried out using an Axygen Maxygen thermocycler (Corning, New York, NY, USA). Amplification of the selected DNA fragment was carried out in a total volume of 25 μL, with 12.5 μL Green GoTaq Master mix (Promega), 2 μL of each primer (10 μM), 3 μL of DNA template, and 7.5 μL of nuclease-free water. The PCR cycling conditions were as follows: an initial denaturing step of 5 min at 94°C, followed by 35 cycles of 92°C for 30 s, 47°C for 45 s, and 72°C for 10 min, and a final extension step at 72°C for 10 min. The PCR products were analysed by electrophoresis in 1% agarose gel using TAE 1X buffer and observed under UV light using the QIAxcel®Advanced System (Qiagen, Hilden, Germany). Purification and sequencing of the PCR products were carried out by Genewiz (South Plainfield, NJ, USA (https://www.genewiz.com/, last accessed July 2023). The barcoding sequences were deposited in GenBank (https://www.ncbi.nlm.nih.gov/genbank/) (accession numbers OR826956-OR826980; Table 1).
Molecular data and phylogenetic analyses
To obtain the consensus sequences of specimens of the species of Serrasentis and G. lepidus, chromatograms of forward and reverse sequences were assembled and edited using the Geneious Pro v. 5.1.7 platform (Drummond et al. Reference Drummond, Ashton, Buxton, Cheung, Cooper, Heled, Kearse, Moir, Stones-Havas, Sturrock, Thierer and Wilson2010). To infer the position and determine sister-group relations of both taxa within a phylogenetic framework of Acanthocephala, the barcoding sequence data generated herein were aligned together with published sequences from GenBank. The sequences included representatives of different families of the order Echinorhynchida and Polymorphida, following the dataset and classification of Verweyen et al. (Reference Verweyen, Klimpel and Palm2011), Gregori et al. (Reference Gregori, Aznar, Abollo, Roura, González and Pascual2013), Barton et al. (Reference Barton, Smales and Morgan2018), Amin et al. Reference Amin, Heckmann, Dallarés, Constenla and Ha2019, Lisitsyna et al. (Reference Lisitsyna, Kudlai, Cribb and Smit2019a), Huston et al. (Reference Huston, Cribb and Smales2020a, Reference Huston, Cribb and Smalesb), Sharifdini et al. (Reference Sharifdini, Amin and Heckmann2020), and Huston and Smales (Reference Huston and Smales2021) (see Online supplementary Table S2 for more details). Adineta gracilis Janson, 1893 and Rotaria rotatoria (Pallas, 1766) were used as outgroups for the phylogenetic analyses in this study, based on their previously established close phylogenetic relationship with Acanthocephala (e.g., Garey et al. Reference Garey, Near, Nonnemacher and Nadler1996; García-Varela et al. Reference García-Varela, Pérez-Ponce de León, de La Torre, Cummings, Sarma and Laclette2000; Huston et al. Reference Huston and Smales2020).
All sequences were aligned using an interface available with MAFFT v. 7.263 (Katoh and Standley Reference Katoh and Standley2016), an “auto” strategy and a gap-opening penalty of 1.53 with Geneious Pro, and a final edition by eye in the same platform. The barcoding sequence dataset was checked, and their nucleotides aligned and examined for the presence of pseudogenes in Geneious Pro, using the translated amino acid sequences based on the invertebrate mitochondrial genetic code. The best partitioning scheme and substitution model for each DNA partition was chosen under the Bayesian Information Criterion (BIC) (Schwarz Reference Schwarz1978) using the ‘greedy’ search strategy in Partition Finder v. 2.1.1 (Lanfear et al. Reference Lanfear, Frandsen, Wright, Senfeld and Calcott2017). The barcoding fragment dataset was partitioned into first-, second- and third-codon positions with the appropriate nucleotide substitution model implemented for each codon position (GTR+I+G for the first [Tavaré Reference Tavaré1986)]; TVM+I+G for the second [Posada, Reference Posada, Baxevanis, Davison, Page, Petsko, Stein and Stormo2003]; and TRN+G for the third codon position [Tamura & Nei, Reference Tamura and Nei1993]).
The barcoding dataset was analyzed with Bayesian inference (BI) through the CIPRES Science Gateway v. 3.3 (Miller et al. Reference Miller, Pfeiffer and Schwartz2010), and by a maximum likelihood (ML) analysis conducted with IQ-TREE v. 1.6.12 (Nguyen et al. Reference Nguyen, Schmidt, von Haeseler and Minh2015). The BI was carried out with MrBayes v. 3.2.2 (Ronquist et al. Reference Ronquist, Teslenko, van der Mark, Ayres, Darling, Höhna, Larget, Liu, Suchard and Huelsenbeck2012). Bayesian and ML inferences were analyzed using a data set partitioned by codon position, and the same models of nucleotide substitution detected by Partition Finder were applied. The Bayesian phylogenetic tree was reconstructed using two parallel runs of Metropolis-Coupled Markov Chain Monte Carlo (MCMCMC) simulations for 20 x 106 generations each. Topologies were sampled every 1,000 generations, and the average standard deviation of split frequencies was observed until it reached < 0.01, as suggested by Ronquist et al. (Reference Ronquist, Teslenko, van der Mark, Ayres, Darling, Höhna, Larget, Liu, Suchard and Huelsenbeck2012). A consensus tree with branch lengths was obtained for the two runs after discarding the first 5,000 sampled trees as burn-in. Support for nodes in BI was evaluated by posterior probabilities (PP). Support for nodes in ML was evaluated with 10,000 ultrafast bootstrap (UFBoot2) approximations (Hoang et al. Reference Hoang, Chernomor, von Haeseler, Minh and Vinh2018) in IQ-TREE. Posterior probability values ≥ 0.95 and UFBoot2 values ≥ 75%, respectively, were considered as strongly supportive of a particular clade. FigTree v. 1.4.4 (Rambaut Reference Rambaut2018) and Adobe Illustrator CS6 were used to visualize and draw BI and ML trees.
Genetic variation parameter estimation
Haplotypes for the barcoding fragment were obtained for 20 adults of the Serrasentis sp. collected from 8 sampling sites (oceanic sampling sites 1–8; Table 1), together with one sequence (GenBank accession number MK937567) previously reported as ‘Serrasentis sagittifer’ from S. papillosum from the continental shelf of the Yucatan Peninsula, Mexico (Vidal-Martínez et al. Reference Vidal-Martínez, Velázquez-Abunader, Centeno-Chalé, May-Tec, Soler-Jiménez, Pech, Mariño-Tapia, Enriquez, Zapata-Pérez, Herrera-Silveira, Hernández-Mena, Herzka, Ordoñez-López and Leopoldina Aguirre-Macedo2019). To assess the completeness of sampling, a haplotype accumulative curve was obtained (Brown et al. Reference Brown, Collins and Boyer2012; Coeur d’acier et al. Reference Coeur d’acier, Cruaud, Artigo, Genson, Clamens, Pierre, Hudaverdian, Simon, Jousselin and Rasplus2014). The genetic variation of the Serrasentis samples studied here was calculated based on the number of haplotypes (h), haplotype diversity (H), and nucleotide diversity (p) (Nei Reference Nei1987), using DnaSP v. 6.12.01 (Rozas et al. Reference Rozas, Ferrer-Mata, Sánchez-Del Barrio, Guirao-Rico, Librado, Ramos-Onsins and Sánchez-Gracia2017). The proportion (p) of absolute nucleotide sites (p-distance) (Nei & Kumar Reference Nei and Kumar2000) was obtained to compare the intra- and interspecific genetic distance of Serrasentis (S. nadakali, S. sagittifer, and the Serrasentis samples from this study) and G. lepidus. The p value matrix was obtained using MEGA v. 11 (Tamura et al. Reference Tamura, Stecher and Kumar2021), with variance estimation, with the bootstrap method (500 replicates) and a uniform nucleotide substitution (transition + transversions) rate.
Results
Serrasentis gibsoni n. sp
General. Medium sized acanthocephalans. Males and females slightly similar in size. Body elongated, cylindrical and pseudo-segmented, presenting a proboscis, neck and trunk (Figures 2a, 3a). Proboscis, anterior, elongate ovoid, armed with 24 longitudinal rows (Figure 5) of 18 hooks in each row (Figure 4); hooks 1–3 long, thin and slightly curved without roots; hooks 4–13 curved and thick, decreasing in length posteriorly, roots robust; hooks 14–18 small and thin with simple roots; ventral hooks more robust than dorsal hooks. Sensory papilla, 1, small circular, on anterior or middle part of the proboscis (Figures 2b, 3b). Neck short, conical and aspinose. Trunk long and cylindrical. Anterior end of the trunk armed with cuticular spines and posteriorly broadened (Figure 4d). Ventral and lateral surface of trunk armed with multiple spinous cuticular combs, characteristic of the genus (Figures 2a, 3a, and 4c). Mature combs consist of spines extend posteriorly in rings along free edge. Proboscis receptacle double walled; cephalic ganglion at about midlevel. Lemnisci long and tubular, approximately twice as long as receptacle. Genital pore subterminal in both sexes.

Figure 2. Serrasentis gibsoni n. sp. (a–e) Male (holotype) from Syacium papillosum; (a) Entire male in ventral view; (b) Proboscis hooks of male; (c) Hooks of ventral longitudinal row; (d) Hooks of dorsal longitudinal row; (e) Male reproductive system. Abbreviations in two lowercase letters: at, anterior testis; bc, bursa copulatrix; cg, cement glands; ci, cirrus; co, comb; le, lemnisci; ne, neck; pa, sensory papilla; po, proboscis receptacle; pr, proboscis; pt, posterior testis; rc, receptacle of cement glands; sp, Saefftigen’s pouch; sv, seminal vesicle; st, spines trunk.
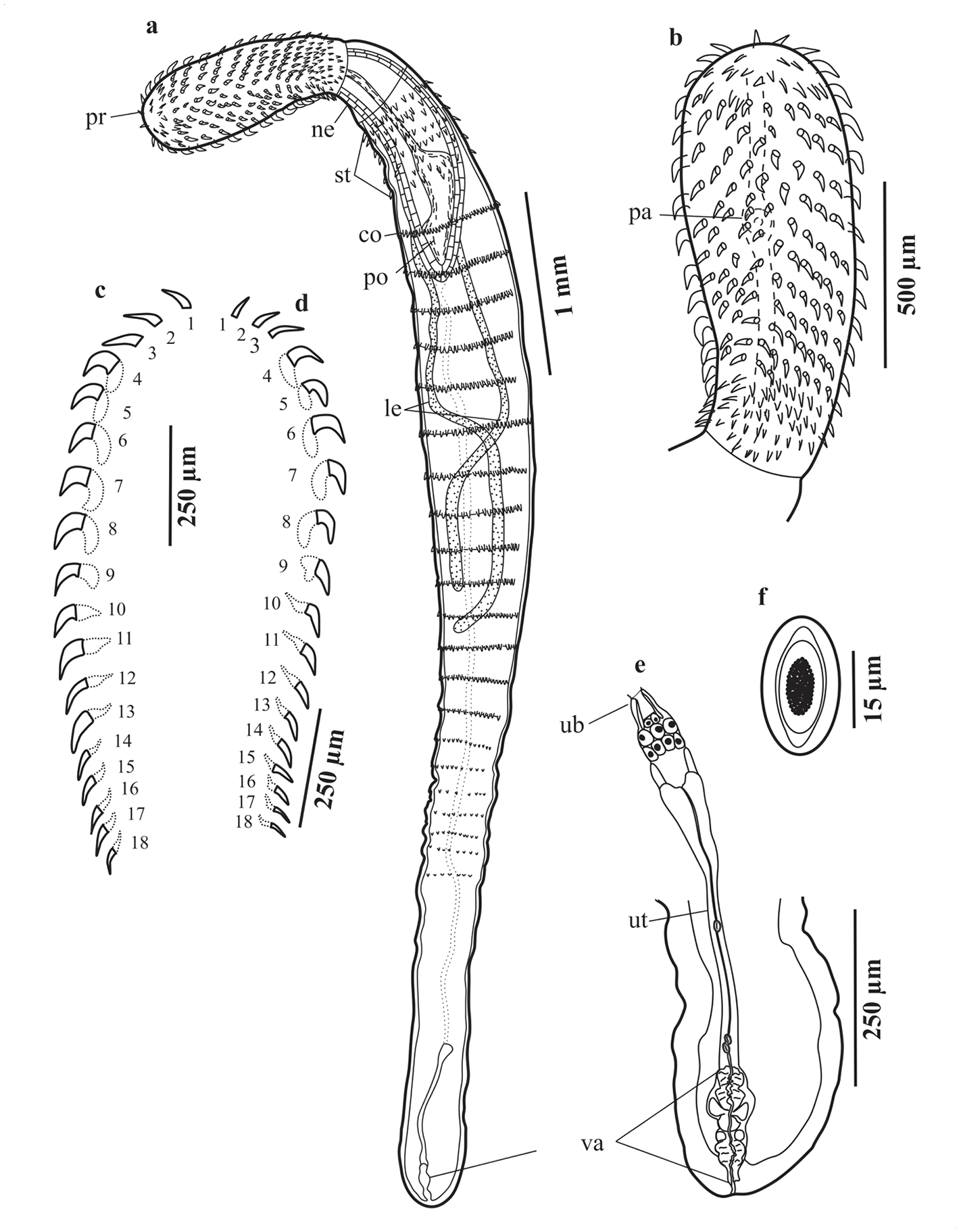
Figure 3. Serrasentis gibsoni n. sp. (a–f) Female (paratype) from Cyclopsetta chittendeni; (a) Entire female in ventral view; (b) Proboscis hooks of female; (c) Hooks of ventral longitudinal row; (d) Hooks of dorsal longitudinal row; (e) Female reproductive system; (f) Egg. Abbreviations in two lowercase letters: co, comb; le, lemnisci; ne, neck; pa, sensory papilla; po, proboscis receptacle; pr, proboscis; st, spines trunk; ub, uterine bell; ut, uterus; va, vagina.

Figure 4. Scanning electron microscopy (SEM) images of Serrasentis gibsoni n. sp. (from two specimens collected from Cyclopsetta chittendeni at two oceanic sampling sites [OSS] from Gulf of Mexico). (a) Proboscis with neck and trunk, male; (b) A magnification of the proboscis; (c) An anterior portion of the trunk showing spines of spine combs (OSS = #5) (d) Spines of the anterior part of trunk showing the anterior and posterior zones without spines, male (OSS = #7). Oceanic sampling site numbers correspond with the numbers in Table 1. Scale bars: 100 μm.

Figure 5. Microphotographs of the apical view of the proboscis of Serrasentis gibsoni n. sp., showing the 24 longitudinal rows. (a) Male collected from Cyclopsetta chittendeni at oceanic sampling site #5 from Gulf of Mexico; (b) Male collected from Syacium papillosum at oceanic sampling site #8 from Gulf of Mexico. Scale bars: 200 μm.
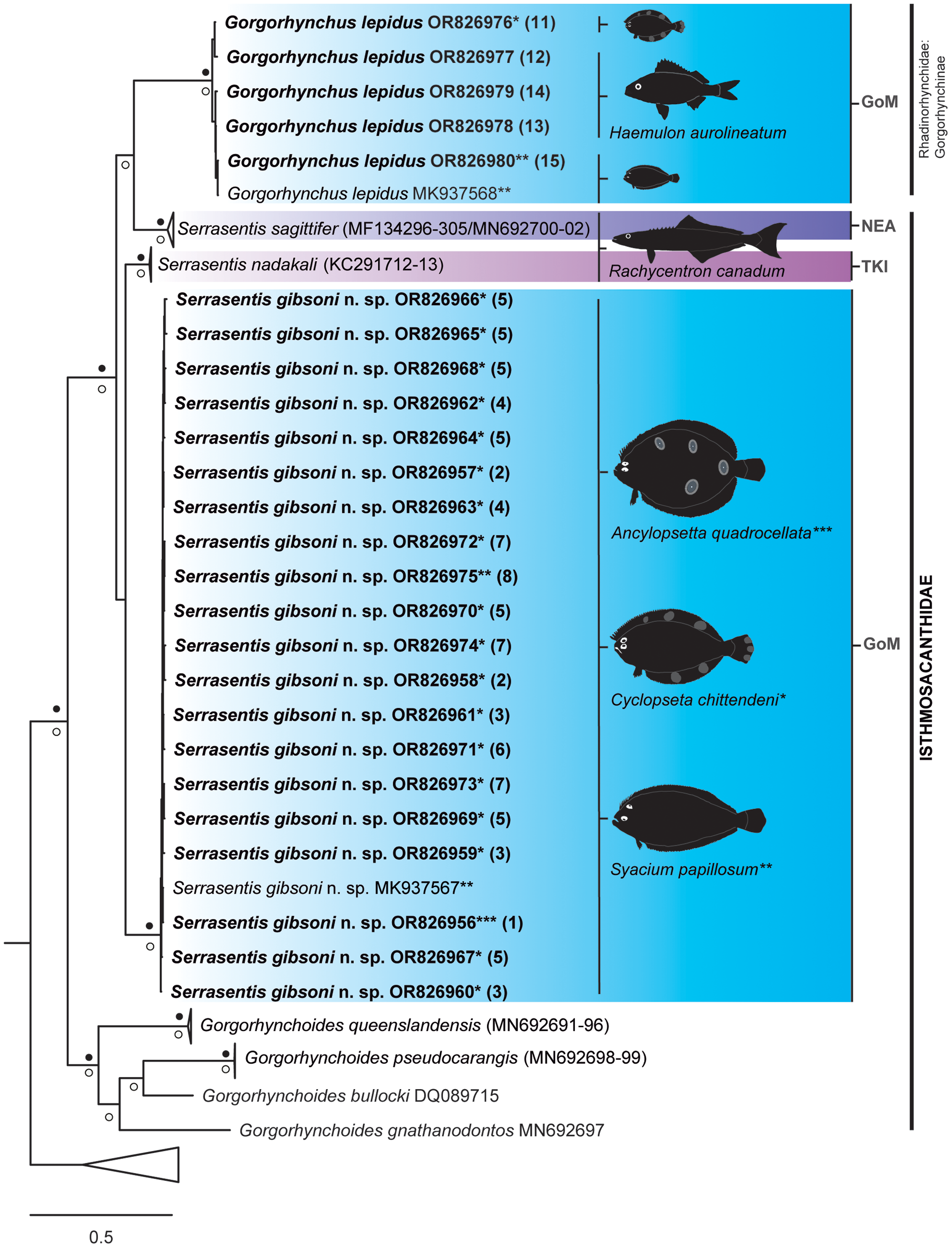
Figure 6. Strict consensus tree resulting from Bayesian inference phylogenetic analysis of barcoding sequence data showing phylogenetic placement of adult acanthocephalan specimens of Serrasentis gibsoni n. sp. and Gorgorhynchus lepidus from the Gulf of Mexico (in bold) relative to other members of the Isthmosacanthidae. Numbers in parentheses following species barcode number/names are cross-referenced in Table 1 and Figure 1. The asterisks (*, **, and ***) refer to species of host where each acanthocephalan sequence was obtained in this study. Filled black circles above and white circles below the branches represent Bayesian posterior probability ≥ 0.95 and maximum likelihood ultrafast bootstrap support values ≥ 75%, respectively. Branch length scale bar at lower left indicates nucleotide substitutions per site. Three letter codes indicate the locality where acanthocephalans were recorded: GoM, Gulf of Mexico, Mexico; NEA, Northeast, Australia; TKI, Trivandrum, Kerala, India.

Figure 7. Haplotype accumulation curve for Serrasentis gibsoni n. sp. in the Gulf of Mexico.
Males (Figures 2a–e). Based on 14 stained and mounted specimens. Trunk 3,375–5,875 (4,962) long, maximum width at level of the middle of trunk 375–700 (551) (Figure 2a). Proboscis 830–1,250 (1,028) long, 240–440 (346) wide (Figure 2b). Serial hook lengths, 2 ventral, 2 dorsal rows measured from anterior: ventral, 60, 37.5; 70, 50; 70, 50; 65, 52.5; 70, 62.5; 70, 60; 70, 50; 65, 57.5; 65, 52.5; 70, 62.5; 70, 75; 65, 62.5; 65, 62.5; 65, 50; 60, 52.5; 50, 50; 50, 45; 50, 30; dorsal, 70, 50; 60, 55; 65, 62.5; 60, 67.5; 55, 65; 60, 62.5; 50, 50; 50, 50; 55, 55; 60, 55; 50, 55; 75, 50; 50, 50; 50, 45; 50, 37.5; 40, 30. Serial root lengths, 2 ventral, 2 dorsal rows measured from anterior: ventral, −; −; −; 40, 62.5; 75, 67.5; 75, 55; 70, 45; 62.5, 45; 62.5, 40; 62.5, 37.5; 62.5, 35; 32.5, 30; 50, 32.5; 45, 30; 37.5, 30; 37.5, 25; 25, 25; dorsal, −; −; −; 30, 50; 50, 62.5; 62.5, 62.5; 62.5, 62.5; 50, 50; 62.5, 45; 55, 45; 55, 50; 50, 47.5; 55, 42.5; 45, 30; 40, 30; 45, 25; 37.5, 25 (Figure 2c–d). Neck 210–430 (329) long, 200–375 (298) wide. Proboscis receptacle 780–1,730 (1413) long, 200–650 (337) wide. Lemnisci 2, 2,100–3,700 (2,663) long, 30–125 (66) wide. Trunk spines anterior, arranged in 11–13 (12) rows and 6–8 (7) circles of spines. Combs transverse ventral 16–20 (17), longest comb spines 50–60 (54). Testes elongated, elliptical, equatorial, between combs 6–12; anterior testes 100–135 (114) long, 25–100 (66) wide; posterior testes 100–150 (107) long, 30–100 (67) wide. Cement glands four elongate, narrow and tubular, 1,075–2,330 (1,687) long, 5–15 (9) wide. Receptacle of cement glands 75–225 (147) long. Saefftigen´s pouch cylindrical 225–520 (378) long. Seminal vesicle 50–180 (124) long. Cirrus 75–275 (117) long. Bursa copulatrix 200–410 (304) long, 85–240 (174) wide (Figure 2e).
Females (Figures 3a–f). Based on 10 stained and mounted specimens. Trunk 2,470–6,200 (4,219) long, maximum width 300–600 (474). Proboscis 650–1,250 (923) long, 260–500 (350) wide. Serial hook lengths, 2 ventral, 2 dorsal rows measured from anterior: ventral, 60, 37.5; 60, 40; 60, 55; 75, 55; 75, 57.5; 75, 55; 75, 52.5; 75, 60; 75, 62.5; 75, 55; 75, 52.5; 85, 55; 75, 50; 75, 40; 50, 40; 50, 37.5; 50, 35; 50, 35; dorsal, 5, 52.5; 75, 47.5; 75, 50; 75, 55; 75, 62.5; 80, 62.5; 75, 60; 75, 52.5; 75, 52.5; 70, 55; 70, 50; 60, 42.5; 75, 47.5; 50, 50; 50, 50; 45, 37.5; 45, 50; 45, 27.5. Serial root lengths, 2 ventral, 2 dorsal rows measured from anterior: ventral, −; −; −; 65, 50;65, 55; 45, 50; 47.5, 37.5; 47, 50; 40, 50; 40, 37.5; 35, 30; 37.5, 35; 35, 25; 45, 30; 45, 27.5; 10, 27.5; 10, 25; 10, 22.5; dorsal, −; −; −; 62.5, 42.5; 62.5, 45; 65, 45; 62.5, 47.5; 55, 35; 50, 20; 30, 25; 37.5, 37.5; 35, 32.5; 30, 35; 27.5, 37.5; 27.5, 30; 25, 25; 20, 25; 20, 20 (Figures 3c–d). Neck 100–380 (255) long, 200–390 (279) wide. Proboscis receptacle 650–1,470 (1,178) long, 180–380 (255) wide. Lemnisci 1,950–3,625 (2,620) long, 40–100 (62) wide. Anterior trunk spines, arranged in 10–14 (12) rows and 5–7 (6) circles. Combs transverse ventral 17–19 (18), longest comb spines 25–60 (46). Female reproductive system 490–915 (761), uterine bell 80–140 (103) long, 30–65 (44) wide. Uterus 225–640 (467) long, 15–40 (30) wide. Vagina 125–190 (164) long, 50–85 (65) wide (Figure 3e). Eggs elliptical, 15–35 (23) long, 10 wide (Figure 3f).
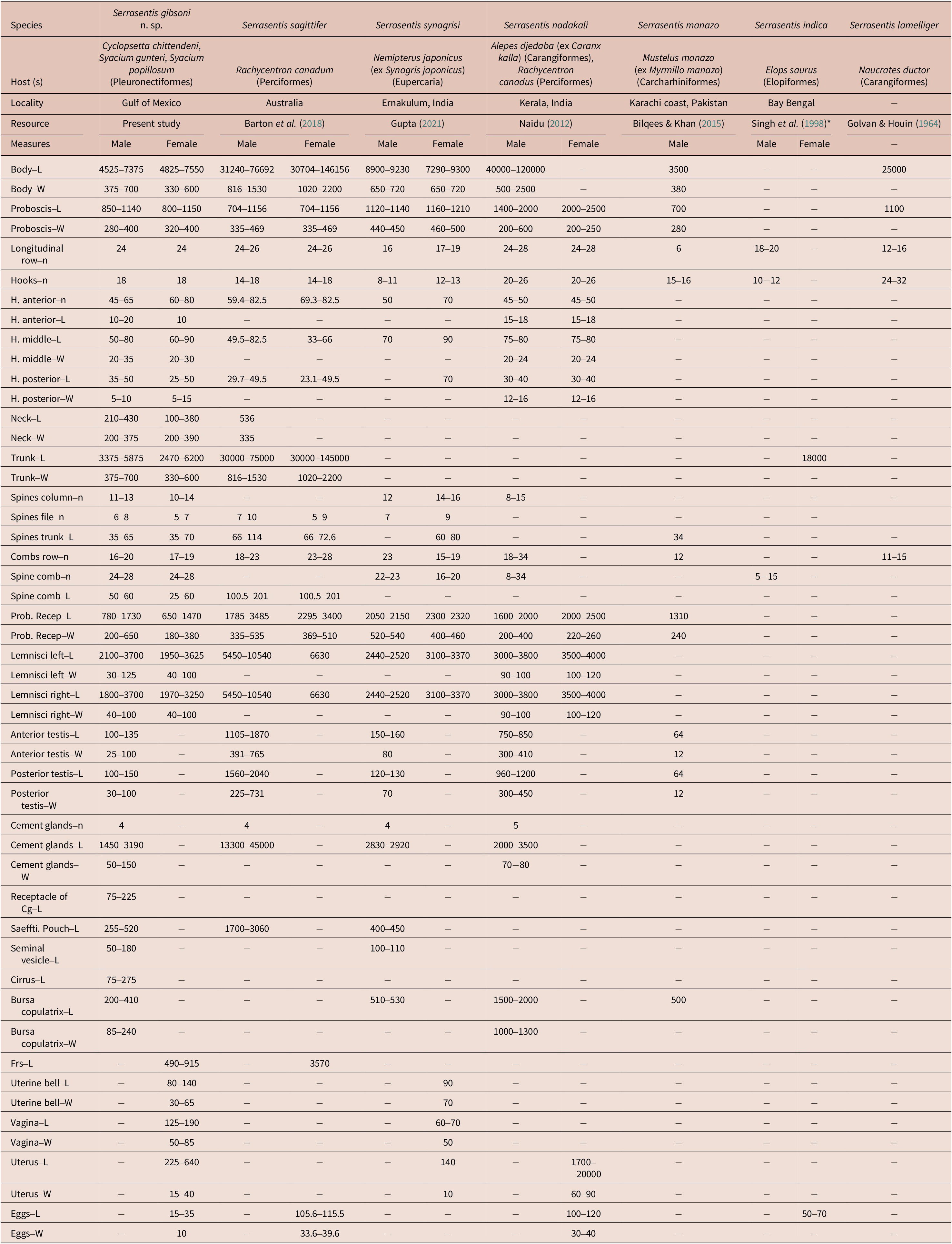
Comparative morphometrics of Serrasentis gibsoni n. sp. with five congeners Serrasentis spp. are given in Table 2. The morphometric measurements of Serrasentis gibsoni n. sp. specimens by host are presented in Online supplementary Table S2.
Table 2. Comparative morphometrics of Serrasentis gibsoni n. sp. with six congeners Serrasentis spp. Measurements in micrometers (μm). Cg, cement gland; Frs, female reproductive system; H, hooks; L, length; N/n, number, Number of examined specimens; Prob. Recep, proboscis receptacle; Saeffti. Pouch, Saefftigen´s pouch; W, weight. The asterisk (*) refers to the publication that contains the description and figures of this species, which could not be procured; therefore, the measurements mentioned for S. indica here are according to Barton et al. (Reference Barton, Smales and Morgan2018) (also see Naidu et al. 2012)
Taxonomic summary
Order Polymorphida Petrochenko, 1956
Isthmosacanthidae Smales, Reference Smales2012
Serrasentis Van Cleave, 1923
Type species: Serrasentis sagittifer (Linton, 1889) Van Cleave, 1932 (type by subsequent designation).
Serrasentis gibsoni n. sp.
Type-host: Dusky flounder, Syacium papillosum (Linnaeus, 1758) (Pleuronectiformes: Paralichthyidae).
Other hosts: Gulf of Mexico ocellated flounder, Ancyclopsetta quadrocellata Gill, 1864; Mexican flounder, Cyclopsetta chittendeni Bean, 1895; Shoal flounder, Syacium gunteri Ginsburg, 1933; (Pleuronectiformes: Paralichthyidae).
Site in infection: Intestine.
Type-locality: Oceanic sampling site #8 (23.31º N; -87.62º W).
Material examined: Holotype, male (CHCM 685); allotype, female (CHCM 683); paratypes, five males, three females (CHCM: 682, 683.2, 684, 685.2, 685.3, 685.4, 686,687).
ZooBank registration: The Life Science Identifier (LSID) for Serrasentis gibsoni n. sp. is: urn:lsid:zoobank.org:pub:55F95417-FA1A-48D9-AD38-90050DAF6EFB
Etymology: This species is named in honor of Dr. David Ian Gibson, in recognition of his outstanding contribution to the knowledge of the field of Parasitology.
Remarks
Due to morphological characteristics such as the presence of multiple spinous combs arranged on irregular transverse rows on the ventrolateral surface, cylindrical proboscis with papillae, and equatorial testes with four cement glands, we consider that this species belongs in the genus Serrasentis Van Cleave, 1923 (see Naidu Reference Naidu2012; Barton et al. Reference Barton, Smales and Morgan2018). Currently, the genus Serrasentis contains six valid species (Barton et al. Reference Barton, Smales and Morgan2018; WoRMS 2023), and within that genus each species can be differentiated by morphological features, especially by the number of longitudinal rows of hooks, the number of hooks in each row, and the number of comb spines present on the trunk. Two species differ from our specimens due to these characteristics: S. indica and S. manazo. Serrasentis indica was recorded and described in the Ladyfish Elops saurus Linnaeus, 1766 from Andhra Pradesh, India; the proboscis of this species is armed with 18–20 longitudinal rows with 10–12 hooks each, and 5–15 comb spines on the ventral trunk (Barton et al. Reference Barton, Smales and Morgan2018). Serrasentis manazo has been reported in starspotted smooth-hound Mustelus manazo Bleeker, 1854 (syn. Myrmillo manazo) from Karachi coast, Pakistan (Bilqees and Khan Reference Bilqees and Khan2015). Serrasentis manazo was described with the proboscis armed with 6 longitudinal rows each with 15–16 hooks, and the posterior part trunk presents with 3 small spines, while S. gibsoni n. sp. has a proboscis armed with 24 longitudinal rows with 18 hooks, trunk with 16–20 (18) comb spines, and the posterior part without spines.
Four other species of Serrasentis presented a range in the number of longitudinal rows of hooks that resembles the number of longitudinal rows of our species. However, S. gibsoni n. sp. differs in other characteristics as follows: Serrasentis sagittifer has a large body size (males = 31–76 mm; females = 30–46 mm); proboscis with 22–24 longitudinal rows with 14–18 hooks each; and the length of the cement gland (maximum sized) representing 58% of the total length of body (Barton et al. Reference Barton, Smales and Morgan2018). Serrasentis nadakali are larger worms, with males measuring up to 120 mm and females up to 170 mm; the proboscis has 24–28 longitudinal rows with 20–26 hooks each, and smaller cement glands, with their length corresponding to 3% of the total length of body (Naidu Reference Naidu2012). Serrasentis laminiger has a medium-sized body (25 mm), the proboscis armed with 31–34 longitudinal rows but with 3–6 hooks each, and maximum spines comb of 16 (Golvan and Houin Reference Golvan and Houin1964). Serrasentis synagrisi has a small body size (males = 8.9–9.2 mm; females = 7.2–9.3 mm); proboscis armed with 16–19 longitudinal rows with 8–13 hooks each; trunk spines anterior, arranged in 12–16 columns and 7–9 rows of spines; combs transverse ventral 15–23 (Gupta Reference Gupta2021). In contrast, S. gibsoni n. sp. differs from these species by presenting a small body (males = 4.5–7.4 mm; females = 4.8–7.5 mm); proboscis armed with 24 longitudinal rows with 18 hooks each; anterior trunk spines in 10–14 (12) rows and 5–8 (7) circles of spines; transverse ventral combs, 16–20 (17); and cement gland medium sized representing 32% of the total length of body.
DNA sequences and dataset analyses
In total, 50 bidirectional barcoding sequences were obtained from 20 adults from Serrasentis gibsoni n. sp. and five adults from Gorgorhynchus lepidus. The barcoding sequences obtained from specimens of S. gibsoni n. sp. had final lengths (in number of base-pairs [bp]) of 669 bp (for 15 sequences), 668 bp (for three sequences), and 660 bp (for two sequences), with a genetic distance of 0.57% between the new mitochondrial sequences together with the sequence MK937567. The length of barcoding sequences from the G. lepidus were 669 (for one sequence), 666 bp (for one sequence), and 663 bp (for three sequences), with a genetic distance of 1.41% between the new barcoding sequences together with the sequence MK937568 previously identified as G. lepidus from S. papillosum from GoM (Vidal-Martínez et al. Reference Vidal-Martínez, Velázquez-Abunader, Centeno-Chalé, May-Tec, Soler-Jiménez, Pech, Mariño-Tapia, Enriquez, Zapata-Pérez, Herrera-Silveira, Hernández-Mena, Herzka, Ordoñez-López and Leopoldina Aguirre-Macedo2019). The total alignment length following the translated amino acid sequences was 678 bp. Nucleotide sequence variation in the barcoding alignment from Echinorhynchida and Polymorphida (excluding the two outgroups) included 201 conserved sites, 471 variable sites, 433 parsimony-informative sites, and 38 singleton sites.
Phylogenetic relationships were inferred using the alignment of the barcoding region, which included 103 sequences from 57 taxa. The phylogenetic trees based on BI and ML analyses were congruent, with differences only affecting non-supported nodes (for more detail, see online supplementary Figures S1 and S2). Following the recommendations of Huston and Smales (Reference Huston and Smales2020), to avoid ambiguities of interpretation about phylogenetical relationships within Acanthocephala due to the molecular evolutionary nature of the barcoding region in this phylum (i.e., rapid evolutionary rate), we focused our results within the clade detected as Isthmosacanthidae + G. lepidus (Rhadinorhynchidae: Gorgorhynchinae), since our study samples (i.e., Serrasentis gibsoni n. sp. and G. lepidus) were phylogenetically located there (Figure 6). In this context, we show the results obtained for this clade in the next section.
Phylogenetic reconstruction and intraspecific variation from barcoding
The twenty-five sequences of acanthocephalan specimens from this study were placed in a greater Isthmosacanthidae + Rhadinorhynchidae (Gorgorhynchinae) clade (PP ≥ 0.95 and UFBoot2 ≥ 75%) and more specifically, grouped with other Serrasentis samples from the GoM, India, and Australia (Figure 6). The clade Isthmosacanthidae + Rhadinorhynchidae (Gorgorhynchinae) comprised 35 sequences (seven species) of Isthmosacanthidae that were found in six host families of marine teleost fishes from Australian waters and the GoM as follows: Gorgorhynchoides lepidus Cable and Mafarachisi, 1970 from Gerreidae fishes; Gorgorhynchoides gnathanodontos Smales, 2014, Gorgorhynchoides pseudocarangis Huston and Smales, 2021 and Gorgorhynchoides queenslandensis Smales, 2014 from Carangidae fishes (for details see Huston et al. Reference Huston, Cribb and Smales2020a, Reference Huston, Cribb and Smalesb); Serrasentis sagittifer from Lutjanidae and Rachycentridae fishes (for details see Barton et al. Reference Barton, Smales and Morgan2018); and Serrasentis gibsoni n. sp. from Paralichthyidae fishes (present study). Six sequences of Gorgorhynchus epidus (Rhadinorhynchidae: Gorgorhynchinae) correspond to Paralichthyidae and Haemulidae fishes from GoM (present study) (for details see Table 1).
Based on the analysis of the barcoding alignment, the sequences generated in this study of S. gibsoni n. sp. from A. quadrocellata, C. chittendeni, and S. papillosum form a well-supported (PP ≥ 0.95 and UFBoot2 ≥ 75%) monophyletic group together with the sequence MK937567. This clade, representing S. gibsoni n. sp., is sister to S. nadakali, but without nodal support. The barcoding tree also shows that all sequences of G. lepidus from C. chittendeni, H. aurolineatum, and S. papillosum form a clade with the sequence MK937568. The clade of G. lepidus is sister to S. sagittifer, but only the ML analyses show high nodal support value (UFBoot2 ≥ 75%).
The genetic distance values of S. gibsoni n. sp., to other Serrasentis species, were 13% and 15.77% for S. nadakali and S. sagittifer, respectively. The genetic distances between S. gibsoni n. sp. and the four congeneric Gorgorhynchoides spp. show a range from 19.53 to 23.93%, while genetic distances of Serrasentis spp. when compared with G. lepidus ranged from 17.72% to 18.33%. The average uncorrected p-distances calculated for the barcoding gene are shown in Table 3. In total, 16 haplotypes among 21 sequences from S. gibsoni n. sp. from the GoM were found, and the haplotype accumulation curve has not yet reached the asymptote (Figure 7). The haplotype diversity (H) was 0.9667 and the nucleotide diversity (π) was 0.0057.
Table 3. Distance matrix of uncorrected p-distances from barcoding sequences among and within pairs of Isthmosacanthidae species (1–7) and rhadinorhynchid Gorgorhynchus lepidus (8) grouped based on Bayesian and maximum likelihood phylogenetic analyses; p-distances are expressed in percentages.

Discussion
In this study, the first Serrasentis species from the intestines of four flatfish species (Paralichthyidae) from the Gulf of Mexico is described, based on morphological and molecular data. Serrasentis gibsoni n. sp. from A. quadrocellata, C. chittendeni, S. gunteri, and S. papillosum, from the Northwest Atlantic Ocean, is the seventh described species of Serrasentis, not including the species with dubious taxonomic descriptions, according to Barton et al. (Reference Barton, Smales and Morgan2018); five valid Serrasentis species, namely S. indica, S. lamelliger, S. manazo, S. nadakali, and S. synagrisi, were described from the Indian subcontinent (Gupta and Jain Reference Gupta and Jain1985; Bhattacharya Reference Bhattacharya2007; Amin Reference Amin2013; Barton and Smales Reference Barton and Smales2015; Amin and Heckmann Reference Amin and Heckmann2021), and a sixth valid species, S. sagittifer with a cosmopolitan distribution, was redescribed from Australian waters (Barton et al. Reference Barton, Smales and Morgan2018).
Five valid species of Serrasentis were reported as adult parasites from four groups of marine teleosts (i.e., Carangiformes, Elopiformes, Eupercaria, Perciformes) (Golvan and Houin Reference Golvan and Houin1964; Naidu Reference Naidu2012; Barton et al. Reference Barton, Smales and Morgan2018; Gupta Reference Gupta2021), and a sixth from the elasmobranch, Mustelus manazo Bleeker, 1854 (ex Myrmillo manazo) (Carcharhiniformes), i.e., S. manazo (Bilqees and Khan Reference Bilqees and Khan2015). Serrasentis gibsoni n. sp. is the first Serrasentis species described from marine teleosts of the order Pleuronectiformes. Barton and Smales (Reference Barton and Smales2015) reported cystacanths of Serrasentis cf. sagittifer from six families belonging to the order Pleuronectiformes (i.e., Bothidae, Citharidae, Cynoglossidae, Paralichthyidae, Psettodidae, and Soleidae). Fonseca et al. (Reference Fonseca, Knoff, Felizardo, Lopes Torres, Nogueira Di Azevedo, Gomes, de São Clemente and Iñiguez2019) reported juvenile specimens of Serrasentis sagittifer from the intestines of two flatfish species (Paralichthyidae), i.e., Paralichthys isosceles Jordan, 1891 and Paralichthys patagonicus Jordan, 1889. Barton and Smales (Reference Barton and Smales2015), Barton et al. (Reference Barton, Smales and Morgan2018), and Fonseca et al. (Reference Fonseca, Knoff, Felizardo, Lopes Torres, Nogueira Di Azevedo, Gomes, de São Clemente and Iñiguez2019) mentioned that the flatfish species are paratenic hosts in the life cycle of S. sagittifer. Amin and Heckmann (Reference Amin and Heckmann2021) reported juveniles and adults of S. sagittifer from eight paratenic (from five fish families, one of which belongs to the family Paralichthyidae) and one definitive host species in the Arabian Gulf, and they discussed their world-wide distribution and comparative morphometrics. In the present study, the four Paralichthyidae flatfish species were definitive hosts of Serrasentis gibsoni n. sp. Kennedy (Reference Kennedy2012) mentioned that crustaceans are intermediate hosts of various acanthocephalan taxa. Barton and Smales (Reference Barton and Smales2015) and Barton et al. (Reference Barton, Smales and Morgan2018) mentioned that the Pleuronectiformes flatfish prefer a diet of crustaceans (e.g., shrimp and crabs). Preferences for crustaceans (e.g., copepods, amphipods, shrimps, and isopods) were also reported for A. quadrocellata, C. chittendeni, S. gunteri, and S. papillosum (Fraser Reference Fraser1971; García-Abad et al. Reference García-Abad, Yánez-Arancibia, Sánchez-Gil and García1992; Sanchez-Gil et al. Reference Sanchez-Gil, Arreguin-Sánchez and García-Abad1994; Kobelkowsky & Rojas-Ruiz Reference Kobelkowsky and Iraiz Rojas-Ruiz2017; Froese & Pauly Reference Froese and Pauly2023). As such, one of the intermediate hosts of S. gibsoni n. sp. is probably a crustacean in the Gulf of Mexico. The host species of Serrasentis gibsoni n. sp. examined in this study are distributed at a depth range of 27–68 m and are demersal or reef-associated in habitat (Froese & Pauly Reference Froese and Pauly2023). Several species of crustaceans from the GoM are sympatric with the definitive host species of Serrasentis gibsoni n. sp. and are found in a similar depth range from demersal habitats, e.g., blue crab (Callinectes sapidus Rathbun, 1896), white shrimp Penaeus setiferus (Linnaeus, 1767), and peneid shrimps (Loesch Reference Loesch1960; Switzer et al. Reference Switzer, Chesney and Baltz2009; Craig Reference Craig2012; Tunell, Reference Tunnell and Ward2017). It is possible that the life cycle of Serrasentis gibsoni n. sp. takes place in demersal and/or reef-associated habitats. Future studies of acanthocephalans from the GoM following a study program as suggested by Blasco-Costa & Poulin (Reference Blasco-Costa and Poulin2017) to elucidate parasite life cycles (i.e., approaches based on morphological matching and/or genetic matching [e.g., Martínez-Aquino et al. Reference Martínez-Aquino, Vidal-Martínez and Aguirre-Macedo2017, Reference Martínez-Aquino, Vidal-Martínez, Ceccarelli, Méndez, Soler-Jiménez and Aguirre-Macedo2020]), can provide a foundation to develop a taxonomical diagnosis of each specific phase of the species of acanthocephalans.
Three acanthocephalan species were previously reported as intestinal and adult parasites of four Pleuronectiformes flatfish species from GoM as follows: 1) Acanthocephaloides plagiusae Santana-Piñeros, Cruz-Quintana, Centeno-Chalé and Vidal-Martínez, Reference Santana-Piñeros, Cruz-Quintana, Centeno-Chalé and Vidal-Martínez2013, described from C. chittendeni and Symphurus plagiusa (Linnaeus, 1766) from the GoM (Santana-Piñeros et al. Reference Santana-Piñeros, Pech and Vidal-Martínez2012, Reference Santana-Piñeros, Cruz-Quintana, Centeno-Chalé and Vidal-Martínez2013; Vidal-Martínez et al. Reference Vidal-Martínez, Velázquez-Abunader, Centeno-Chalé, May-Tec, Soler-Jiménez, Pech, Mariño-Tapia, Enriquez, Zapata-Pérez, Herrera-Silveira, Hernández-Mena, Herzka, Ordoñez-López and Leopoldina Aguirre-Macedo2019; Centeno-Chalé et al. Reference Centeno-Chalé, Aguirre-Macedo, Gold-Bouchot and Vidal-Martínez2015), suggesting a host-specificity pattern; 2) Gorgorynchus lepidus from S. papillosum (Vidal-Martínez et al. Reference Vidal-Martínez, Velázquez-Abunader, Centeno-Chalé, May-Tec, Soler-Jiménez, Pech, Mariño-Tapia, Enriquez, Zapata-Pérez, Herrera-Silveira, Hernández-Mena, Herzka, Ordoñez-López and Leopoldina Aguirre-Macedo2019). In the present study, adult specimens of G. lepidus are recorded for the first time as parasites of the flatfish A. quadrocellata and the tomtate grunt H. aurolineatum; 3) ‘Serrasentis sagittifer’ from C. chittendeni, S. gunteri, and S. papillosum (Vidal-Martínez et al. Reference Vidal-Martínez, Centeno-Chalé, Torres-Irineo, Sánchez-Ávila, Gold-Bouchot and Aguirre-Macedo2014, Reference Vidal-Martínez, Velázquez-Abunader, Centeno-Chalé, May-Tec, Soler-Jiménez, Pech, Mariño-Tapia, Enriquez, Zapata-Pérez, Herrera-Silveira, Hernández-Mena, Herzka, Ordoñez-López and Leopoldina Aguirre-Macedo2019; Centeno-Chalé et al. Reference Centeno-Chalé, Aguirre-Macedo, Gold-Bouchot and Vidal-Martínez2015). Based on the specimens examined in this study and described as Serrasentis gibsoni n. sp. from A. quadrocellata, C. chittendeni, S. gunteri, and S. papillosum from the GoM, we believe that the previously published record of ‘Serrasentis sagittifer’, i.e., from Vidal-Martínez et al. (Reference Vidal-Martínez, Centeno-Chalé, Torres-Irineo, Sánchez-Ávila, Gold-Bouchot and Aguirre-Macedo2014, Reference Vidal-Martínez, Velázquez-Abunader, Centeno-Chalé, May-Tec, Soler-Jiménez, Pech, Mariño-Tapia, Enriquez, Zapata-Pérez, Herrera-Silveira, Hernández-Mena, Herzka, Ordoñez-López and Leopoldina Aguirre-Macedo2019) and Centeno-Chalé et al. (Reference Centeno-Chalé, Aguirre-Macedo, Gold-Bouchot and Vidal-Martínez2015), associated with the same flatfish host species, i.e., C. chittendeni, S. gunteri, and S. papillosum, and from the same oceanic sampling sites analysed in this study, most likely correspond to Serrasentis gibsoni n. sp. Unfortunately, we were unable to obtain the specimens from previous studies to corroborate their taxonomical identity.
Phylogenetic analyses grouped Serrasentis gibsoni n. sp. within the Isthmosacanthidae, and it is related to S. nadakali, as shown in both phylogenetic analyses (IB and ML) but without nodal support (Figure 6, online supplementary Figures S1 and S2). The phylogenetic relationships obtained from the barcoding dataset for taxa of Isthmosacanthidae were similar to those found in previous phylogenetic analyses carried out for similar taxa using the same gene (e.g., Braicovich et al. Reference Braicovich, Lanfranchi, Farber, Marvaldi, Luque and Timi2014; Barton et al. Reference Barton, Smales and Morgan2018; Lisitsyna et al. Reference Lisitsyna, Kudlai, Cribb and Smit2019a; Huston et al. Reference Huston, Cribb and Smales2020a; Sharifdini et al. Reference Sharifdini, Amin and Heckmann2020; Huston & Smales Reference Huston and Smales2021).
The genetic distance analysis supported Serrasentis gibsoni n. sp. as a distinct species from those previously described and that have DNA sequence data available for comparison. Uncorrected p-distance analyses revealed ≥ 13% differences based on the barcoding gene among Serrasentis gibsoni n. sp., S. nadakali, and S. sagittifer, the latter being the one with the highest percentage of divergence (15.77%). These values are comparable to the barcoding genetic distances from other studies (Barton et al. Reference Barton, Smales and Morgan2018). The barcoding sequence data generated for Serrasentis gibsoni n. sp. individuals from eight different oceanic sites in the GoM revealed extremely high intraspecific genetic variation (0.0057 nucleotide diversity). Martínez-Aquino et al. (Reference Martínez-Aquino, Vidal-Martínez, Ceccarelli, Méndez, Soler-Jiménez and Aguirre-Macedo2020) recently detected high intraspecific genetic variation with similar values (0.0086 nucleotide diversity), using the same molecular marker for the trypanorhynch cestode, Oncomegas wageneri (Linton, 1890) Dollfus, 1929, from C. chittendeni from the GoM. However, it is necessary to include more parasites from many more oceanic sites and a wider range of molecular markers to describe how evolutionary forces act within the parasite populations of the GoM (e.g., population genetic structure).
Phylogenetical relationships detected for Serrasentis gibsoni n. sp. as parasites of four species of Paralichthyidae flatfishes may reflect a host-specificity pattern for the Paralichthyidae–Serrasentis gibsoni n. sp. association from the GoM. This specificity pattern can be supported based on the values of prevalence (%) recorded between the host-parasite association for Paralichthyidae–Serrasentis gibsoni n. sp.; e.g., Serrasentis gibsoni n. sp. (ex. ‘S. sagiffiter’) as a parasite of C. chittendeni (= 24%) (Centeno-Chalé et al. Reference Centeno-Chalé, Aguirre-Macedo, Gold-Bouchot and Vidal-Martínez2015), Syacium gunteri (= 4.50%), and Syacium papillosum (= 15.74%) (Vidal-Martínez et al. Reference Vidal-Martínez, Velázquez-Abunader, Centeno-Chalé, May-Tec, Soler-Jiménez, Pech, Mariño-Tapia, Enriquez, Zapata-Pérez, Herrera-Silveira, Hernández-Mena, Herzka, Ordoñez-López and Leopoldina Aguirre-Macedo2019). Centeno-Chalé et al. (Reference Centeno-Chalé, Aguirre-Macedo, Gold-Bouchot and Vidal-Martínez2015) reported that the dynamics of the prevalence values of Serrasentis gibsoni n. sp. (ex. ‘S. sagiffiter’) from C. chittendeni can change over five months from ranges of 8–24% due to impacted and healthy environments, reflecting a parasitic re-population rate for acanthocephalans in the face of environmental disturbances and seasonal fluctuations. This empirical evidence of prevalence variation led Vidal-Martínez (2016, 2019) to suggest that the life cycles of the acanthocephalan species associated with sole are closely linked to both the presence of intermediate and definitive hosts, as well as the environmental conditions in specific marine sites. Similar population fluctuation patterns were reported from A. plagiusae (see Santana-Piñeros et al. Reference Santana-Piñeros, Pech and Vidal-Martínez2012, Reference Santana-Piñeros, Cruz-Quintana, Centeno-Chalé and Vidal-Martínez2013), an acanthocephalan species apparently also specific to flatfish from the GoM. In fact, based on the ecological dynamics of the parasitic infection (e.g., prevalence values), host-specificity pattern observed between Paralichthyidae–Serrasentis gibsoni n. sp., and the high population genetic variation detected for Serrasentis gibsoni n. sp., it is possible that this group of flatfish hosts can be a niche for diversification, at least for this species.
In this study, a rhadinorhynchid species, Gorgorhynchus lepidus, was included for the first time in a phylogeny within Acanthocephala, based on the barcoding gene. Based on our phylogenetical analyses, G. lepidus is sister to S. sagittifer, but only the ML analyses recovered this relationship with high nodal support. Rhadinorhynchidea was recovered as a paraphyletic assemblage (García-Varela & Nadler Reference García-Varela and Nadler2005; Verweyen et al. Reference Verweyen, Klimpel and Palm2011; Gregori et al. Reference Gregori, Aznar, Abollo, Roura, González and Pascual2013; Abdel-Ghaffard et al. Reference Abdel-Ghaffar, Morsy, Abdel-Gaber, Mehlhorn, Al Quraishy and Mohammed2014; Braicovich et al. Reference Braicovich, Lanfranchi, Farber, Marvaldi, Luque and Timi2014; Barton et al. Reference Barton, Smales and Morgan2018; Lisitsyna et al. Reference Lisitsyna, Kudlai, Cribb and Smit2019a; Steinauer et al. Reference Steinauer, Garcia-Vedrenne, Weinstein and Kuris2019; Huston et al. Reference Huston, Cribb and Smales2020a; Sharifdini et al. Reference Sharifdini, Amin and Heckmann2020). Pichelin & Cribb (Reference Pichelin and Cribb2001) mentioned that the number of cement glands is a distinguishing character for the recognition of acanthocephalan families, especially for the Rhadinorhynchidae. Other authors (e.g., Amin et al. Reference Amin, Heckmann and Ha2011; Amin Reference Amin2013) have argued that the number of cement glands is not a useful character to discriminate the genera in the Rhadinorhynchidae, including Rhadinorhynchus (i.e., 2–8 cement glands). Golvanorhynchus Noronha, Fabio & Pinto, 1987, Gorgorhynchoides Cable & Linderoth, 1963, Isthmosacanthus Smales, 2012, and Serrasentis are the four recognized genera in the Isthmosacanthidae (Smales Reference Smales2012; Huston & Smales Reference Huston and Smales2021). Amin (Reference Amin2013) accepted the validity of the Isthmosacanthidae but rejected inclusion of Gorgorhynchoides and Golvanorhynchus, on the basis that the characteristics used by Smales (Reference Smales2012) to unite Golvanorhynchus, Gorgorhynchoides, and Isthmosacanthus, namely six cement glands, a similar proboscis shape, a trunk swelling and trunk spines, were also shared by many other rhadinorhynchids. However, only Serrasentis and Gorgorhynchoides have been tested based on molecular phylogenetical analyses (e.g., Verweyen et al. Reference Verweyen, Klimpel and Palm2011; Gregori et al. Reference Gregori, Aznar, Abollo, Roura, González and Pascual2013; Abdel-Ghaffard et al. 2014; Braicovich et al. Reference Braicovich, Lanfranchi, Farber, Marvaldi, Luque and Timi2014; Barton et al. Reference Barton, Smales and Morgan2018; Lisitsyna et al. Reference Lisitsyna, Kudlai, Cribb and Smit2019a; Steinauer et al. Reference Steinauer, Garcia-Vedrenne, Weinstein and Kuris2019; Huston et al. Reference Huston, Cribb and Smales2020a; Sharifdini et al. Reference Sharifdini, Amin and Heckmann2020; Huston & Smales, Reference Huston and Smales2021; present study). Gorgorhynchoides spp. have six cement glands (Smales Reference Smales2012; Huston & Smales, Reference Huston and Smales2021), whereas the species of Serrasentis and Gorgorhynchus have four cement glands (Pichelin & Cribb Reference Pichelin and Cribb2001; Barton et al. Reference Barton, Smales and Morgan2018; also see Golvan Reference Golvan1969; Salgado-Maldonado et al. Reference Salgado-Maldonado1978; Smales et al. Reference Smales, Barton and Chisholm2019). Huston & Smales (Reference Huston and Smales2021), based on molecular phylogenetical evidence, made an amendment to transfer Gorgorhynchoides spp. (i.e., G. gnathanodontos, G. pseudocarangis, and G. queenslandensis) and Serrasentis (i.e., S. saggitiffer and S. nadakali) to Isthmosacanthidae. In this study, it is interesting to observe that the species of Serrasentis and Gorgorhynchus (i.e., species with four cement glands) form a separate clade to that of the species of Gorgorhynchoides spp. (i.e., species with six cement glands), perhaps because Gorgorhynchoides does not belong in the same group as the other two genera, as proposed by Amin (Reference Amin2013). The number of cement glands may be a synapomorphy at the level of natural groups (e.g., genera and families); however, at higher taxonomic hierarchies, it could be expressed as evolutionary convergence. Future studies of morphological character mapping over molecular phylogenetic trees (e.g., Huelsenbeck et al. Reference Huelsenbeck, Nielsen and Bollback2003, Maddison & FitzJhon Reference Maddison and FitzJohn2015), including more species of Rhadinorhynchidae (e.g., Gorgorhynchus spp.) and Isthmosacanthidae, would be able to confirm or falsify the phylogenetic relationships detected here between Gorgorhynchus and Isthmosacanthidae, and elucidate the evolution of their morphological traits (i.e., number of cement glands).
Supplementary material
The supplementary material for this article can be found at http://doi.org/10.1017/S0022149X23000822.
Acknowledgments
We are grateful to Arturo Rincón Sandoval (Ms. Sc. student, MyDCI, Laboratorio de Biología Evolutiva de Parásitos (BEP-FC-UABC)) for technical support with the digital image editions and Arianna Jimenez Ruiz for the information capture. JGGT was supported by a CONAHCyT scholarship (#637098) to obtain his Ms. Sc. degree. The authors also thank the staff of the laboratory of Patología Acuática: Clara Vivas Rodríguez, Arturo Centeno Chale, Gregory Arjona Torres, and Francisco Puc Itzá from CINVESTAV-IPN, Unidad Mérida, Mexico for collecting the material for this study. They are grateful to MSc. Abril Gamboa for her technical assistance in the molecular techniques. We are grateful to Sean M. Rovito for help with language editing and constructive criticism. The manuscript has greatly benefited from the comments of two anonymous referees.
Financial support
This work was financially supported by the Consejo Nacional de Humanidades, Ciencia y Tecnología (CONAHCyT) (Mexican Ministry of Energy) Hydrocarbon Trust, project #201441 to VMVM and MLAM. This is a contribution of the GoM Research Consortium (CIGoM). Also, this work was partially funded by Grant #A1-S-15134 from the CONAHCyT to FSC, and by Grant #400/1/C/23/23 (23a Convocatoria Interna UABC-2023) to AMA.
Competing interest
None.
Ethical standard
The statement of ethics approval for this study was provided by the Institutional Animal Care and Use Committee (IACUC) from the Center of Research and Advanced Studies (protocol number 0138-15, available upon request).