Introduction
Congenital heart disease is the most common birth defect affecting approximately 0.8% of all births [Reference Reller, Strickland, Riehle-Colarusso, Mahle and Correa1]. Critical congenital heart disease (CCHD) accounts for approximately 20% of congenital heart disease and is life-threatening if not timely diagnosed [Reference Reller, Strickland, Riehle-Colarusso, Mahle and Correa1–Reference Chang, Gurvitz and Rodriguez4]. In fact, prior to universal oxygen saturation-based CCHD screening, 25% of CCHD were diagnosed after hospital discharge, with some diagnosed at autopsy [Reference Wren, Reinhardt and Khawaja2, Reference Lannering, Bartos and Mellander5]. Oxygen saturation screening has since reduced mortality associated with CCHD and helped with earlier diagnosis, but nearly 900 neonates with CCHD remain undiagnosed annually in the USA [Reference Ailes, Gilboa, Honein and Oster6, Reference Abouk, Grosse, Ailes and Oster7]. Coarctation of the aorta (CoA) is the most commonly missed CCHD defect despite oxygen saturation screening as it is associated with poor systemic perfusion without hypoxemia [Reference Lannering, Bartos and Mellander5, Reference Ailes, Gilboa, Honein and Oster6, Reference Peterson8]. Late diagnosis of defects such as CoA can be particularly detrimental. Preliminary analysis of CCHD defects missed by oxygen saturation screening demonstrates that 18% of newborns with late CoA diagnosis die, some before surgery can be done (unpublished data) [Reference Hogan9]. More than 50% of deaths due to missed CCHD occur before corrective surgery, either at home or shortly after arriving to the Emergency Department [Reference Chang, Gurvitz and Rodriguez4].
To improve CCHD detection and minimize false-negative screens, especially of defects associated with poor perfusion such as CoA, researchers have investigated the addition of non-invasive pulse oximetry measurements such as perfusion index (PIx), radiofemoral pulse delay, and other photoplethysmography waveform characteristics to the current oxygen saturation CCHD screening algorithm [Reference Siefkes10–Reference Sorensen, Sadiq, Clifford, Maher and Oster14]. However, accessing and interpreting these additional pulse oximetry data are not a prevalent practice. In addition, pulse oximetry devices currently only store de-identified data, limiting its utility for research. In part due to these limitations, prior studies evaluating PIx have either included brief (10 s) clinician interpreted and manually documented perfusion indices or have utilized retrospective de-identified samplings from pulse oximetry devices from routine screening [Reference Siefkes10–Reference Schena12, Reference Jegatheesan, Nudelman, Goel, Song and Govindaswami15].
To solve this problem, we created an automated real-time data collection and analysis architecture that is able to perform non-invasive measurement of perfusion and oxygenation data. The system is able to wirelessly communicate with pulse oximetry devices and store data on secure computing systems. The main focus of this paper is the novel data collection pipeline that is customizable by providers depending on the target clinical outcomes. To date, there are no other published research or clinical tools capable of prospectively collecting abundant high-fidelity CCHD screening data. The insights driven by this data collection will be beneficial to developing future algorithmic CCHD screening processes and may be applicable to other vascular disease processes that could benefit from non-invasive pulse oximetry perfusion diagnostics (aneurysms, aortic dissections, atherosclerosis, thrombosis, or vascular graft monitoring).
Methods
The primary focus of our overall study was to explore correlations between oxygen saturation and perfusion data in neonates with and without CCHD. The study was registered on ClinicalTrials.gov (NCT04056104). The study was approved by the Institutional Review Boards at all sites. The parents/guardians of all subjects provided informed consent prior to study enrollment. As part of this study, we developed an integrative software and hardware system to collect necessary physiological data. Our system needed to: (1) enable continuous and automated data collection and storage from multiple pulse oximeters simultaneously; (2) ensure accurate time-stamping of the collected data; and (3) allow ease of use by non-technical end users in terms of both data acquisition hardware and data management software.
Defects such as CoA are associated with differences in oxygen saturation and PIx from the right upper extremity (pre-ductal) and any lower extremity (post-ductal). Most current CCHD screening methods employ the sequential application of pulse oximetry probe to the right upper extremity followed by a lower extremity with manual documentation and interpretation of data. Simultaneous collection of data using dual pulse oximetry (Fig. 1) enables accurate collection and interpretation of data using a simple workflow (Fig. 2 and Table 1).
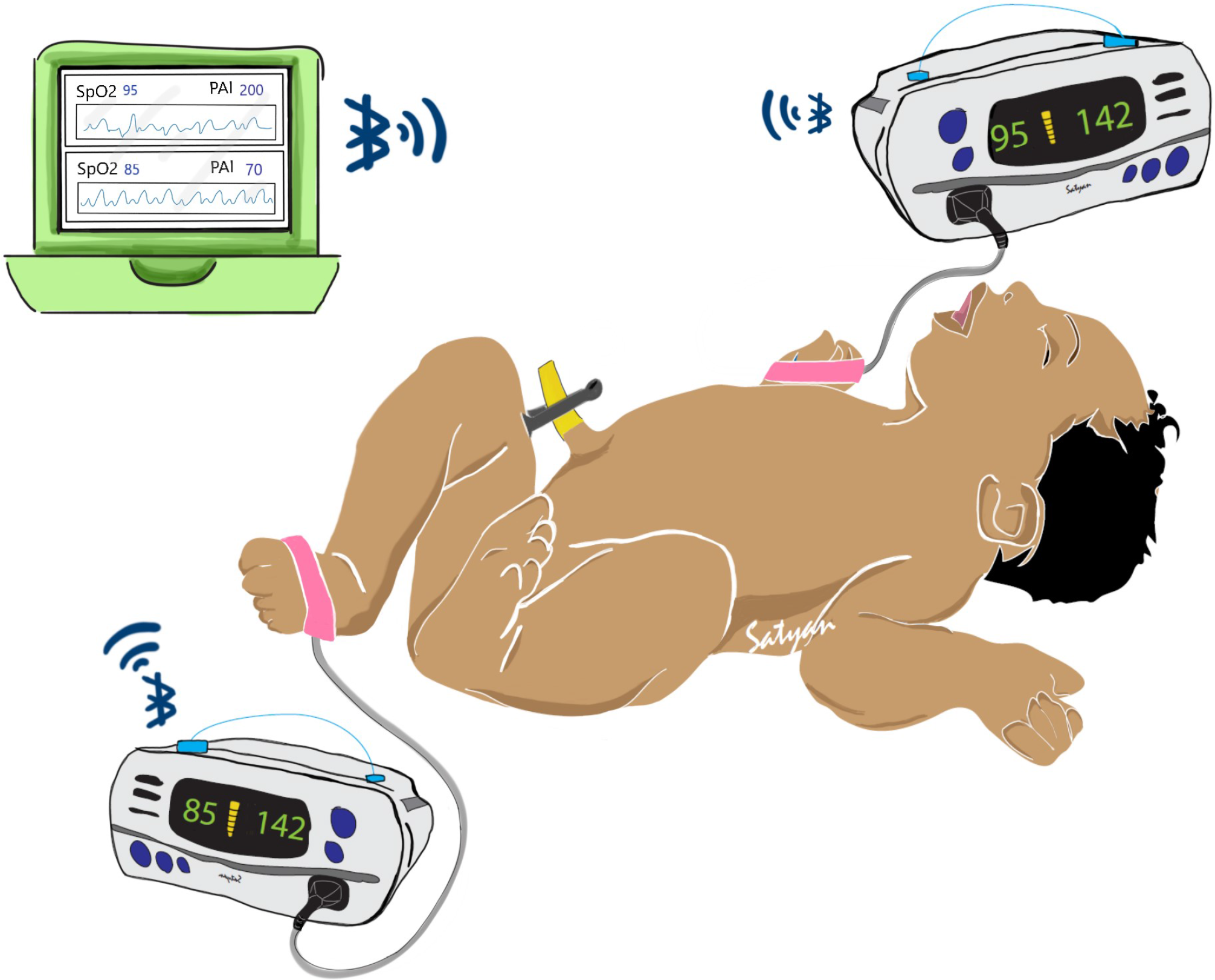
Fig. 1. Illustration of potential uses for this system in post-delivery critical congenital heart disease (CCHD) screening. Two pulse oximeter devices are applied to both a foot and right hand of a neonate. In our case, we use the Nonin® WristOx2™ 3150 to communicate wirelessly with a central aggregator device. The aggregator can then perform visualization and analytics on whether the neonate displays risk for CCHD while simultaneously storing the data for later review.

Fig. 2. Data collection workflow. Illustration of the workflow of the system. A technician controls the software and attaches the pulse oximeters to the patient. They then enter the patient identification number and other medical details. The software will automatically connect to the oximeters via Bluetooth, displays the oximetry and perfusion data in real time, and stores the data.
Table 1. Raw data fields collected by our system

Our general system consists of two independent perfusion and oxygenation monitors (oximeters) that are attached to the study subject’s right hand and any foot, and a central aggregator device that is able to communicate with the oximeters and temporarily store their data for eventual retrieval. For our oximeter, we use the Nonin® WristOx2™ 3150. This device allows simultaneous collection of photoplethysmography and oxygenation and perfusion data, can externally transmit data using Bluetooth and is relatively inexpensive. We chose the Pi-top™, an affordable laptop computer that uses Raspberry Pi microcomputers, as our central aggregator. By developing appropriate software, we were able to collect oxygenation and perfusion data via Bluetooth through the Nonin® devices. The Pi-top is also able to maintain temporal alignment between the two separate Nonin® devices. This ensured that data are accurately time-stamped and synchronized. Time-stamps can also be adjusted to ensure complete de-identification. Our entire system is illustrated in Figure 1.
Once collected and dated, data files are automatically aggregated and coded with unique study identifiers. The data are then transferred from the Pi-top using a secure USB drive and uploaded to REDCap through a secure data transfer mode. This architecture and workflow (Fig. 2) enable ease of data collection and data sharing across different hospitals.
Results
Data Collection
Using this system, we have gathered data from 190 newborns. At time of enrollment, 130 were presumed to be without congenital heart disease and 60 were expected to have congenital heart disease. In total, we have gathered approximately 1665 min of pulse oximetry data from these babies. To date, we have confirmed 60 of the newborns enrolled as presumably healthy newborns remain healthy. One baby enrolled as a presumably healthy baby before qualifying for the routine CCHD screen, was determined to have CCHD based on the first study measurements collected (oxygen saturations were in the 60s). The follow-up is still pending for the remaining babies enrolled as presumably healthy. Of the babies enrolled presumably with congenital heart disease, we have confirmed the final classification for 48 (31 = CCHD, 13 = non-critical CHD, and 4 = without CHD). In total, this comprises 400 min of oximetry data from newborns with the final classification of congenital heart disease (300 min = CCHD, 100 min = non-critical CHD) and 610 min of oximetry data from newborns confirmed to be without congenital heart disease. The majority of patients so far were enrolled at the University of California, Davis, but four other hospitals (Sutter Medical Center, Sacramento, University of California, Los Angeles, University of California, San Francisco, and Cohen’s Medical Center, New York) are participating in the study with plans to enroll hundreds of more newborns over a total 2-year period.
The detailed process of the data collection is as follows: We attach two Nonin® WristOx2™ 3150 pulse oximeters to the subject, one on the right hand and the other on either foot. Once these Nonin® devices display valid measurements, and we can ensure minimal signal noise resulting from limb movement, we initialize our collection software on the Pi-top™ device. In the software user interface, a unique study identification number is entered. This study identifier is an anonymous token that can only be linked back to the patient via an encrypted database like REDCap. Next, we input the fraction of inspired oxygen (FiO2) the patient is receiving. For study protocols that involve repeat measurements, we included an alphanumeric entry to label specific measurements for later identification. The software connects to both pulse oximeters via Bluetooth and starts collecting pulse oximetry, oxygen saturation, heart rate, pulse amplitude index (PAI) – synonymous with PIx, and photoplethysmography data. All data are time-stamped so we can maintain temporal accuracy between the two Nonin® devices. During collection, data are also displayed on the Pi-top screen in real time, which aids with data quality control (shown in Fig. 3). To further demonstrate our process, we provided stepwise instructions in an online supplement.

Fig. 3. Examples of features that can be extracted from raw waveform. Features that can be extracted from raw waveform include pulse amplitude index (PAI) (Box A), heart rate variability (Box A), radiofemoral delay (f-h TD) (Box B), and both the systolic rise and diastolic fall slope of the photoplethysmography waveform (Box C).
Data Aggregation
Management of each Pi-top™, including data aggregation, is performed by a technician or research coordinator present on the site. During aggregation, a human must manually ensure that data is correctly aggregated to the database. The technician (1) uploads data to the REDCap database, (2) backs up the folders in the Pi-top™ to an external hard drive, and (3) deletes all the files on the Pi-top™ when the data are stored locally somewhere. Once these steps are complete, uploaded data can be accessed across the research network and referenced by the patient identifier.
Scalability and Adding New Devices
Provisioning additional Pi-top™ and pulse oximeter devices enable the collection of multiple patients simultaneously and prevent data collection slowdowns in study progress due to lost or damaged equipment. To more rapidly provision devices, we developed an image of the Pi-top™ with the pre-configured details of our software that can be copied to the microSD card of the Pi-top™ in a single step resulting in a fully functional Pi-top™ without the need to install multiple software components. This workflow substantially decreases the number of steps necessary for Pi-top™ deployment and may allow non-technical users to assist more experienced personnel in the provisioning process, which we have successfully done remotely with sites when a software change was needed.
Data Collected and Feature Identification
Table 1 summarizes the data fields collected from the Nonin® devices for each patient enrolled. The data are visualized on the Pi-top (Supplemental Figure) for real-time interpretation and then can be reconstructed from the logged data for analysis and feature detection on our compute server. From the collected data, we are able to extract different features stated in Table 2. For example, Fig. 3B and C illustrates how the delays between systolic peaks of hand and foot waveforms (Feature #6 in Table 2) and the slopes of the systolic rise and diastolic fall (Feature #3 and #4 in Table 2) can be extracted from reconstructed waveforms. Various statistics of these features (minimum, maximum, mean, standard deviation, and median) may be useful for further characterization and development of the prediction model for CCHD detection.
Table 2. Features that can be extracted from the data fields collected

To further demonstrate the features collected in our data, we included photoplethysmography waveforms from a healthy baby (Fig. 4A) and a baby with critical CoA (Fig. 4B and C). Interestingly, the baby with CoA was trialed off prostaglandin E1 therapy to assess the severity of the coarctation and thus the ductus arteriosus was presumably closing and the CoA narrowing during our data collection. Our measurements occurred approximately 10 h and 36 h after the prostaglandin E1 infusion was discontinued. The baby with CoA was monitored in the neonatal intensive care unit and was asymptomatic. The baby had an echocardiogram at 3 days of age, which noted the ductus arteriosus was closed and the CoA was minimal. Thus, the baby was discharged home with plans to follow-up outpatient with cardiology. The baby then represented to care at 7 days of age, and an echocardiogram showed an increase in the gradient across the CoA prompting readmission and subsequent surgery for CoA repair. Note the foot pulse amplitude index (PAI) decreased in this neonate with increasing duration off prostaglandin E1 infusion. The average foot PAI decreased from 96 to 36 during this time period (Fig. 4B and C).

Fig. 4. Example of pulse oximetry data collected from a healthy newborn and a newborn with critical coarctation of the aorta (CoA). Solid lines are from raw data. Dashed lines have a filtered applied to assist with peak identification due to the dicrotic notch interfering with peak identification for the infant with coarctation. (Box A) A normal newborn demonstrates minimal time delay between the right hand and foot pulse (f-h TD) and similar pulse amplitude index (PAI) in hand and foot. (Box B and C) A newborn with critical CoA shows the foot PAI decrease and the f-h TD change as more time off prostaglandin E1 passes. Additionally, the dicrotic notch appearance in the right hand is notable different in the baby with CoA compared to both the healthy newborn and the earlier measurement in the same baby when the ductus arteriosus was presumably more open.
There are some other notable features in this baby with CoA compared to the healthy baby (Fig. 4). Note the closer proximity of the peaks of the hand and foot (f-h TD) in the healthy baby versus the baby with CoA. Additionally, note how the f-h TD changes as more hours off prostaglandin pass and presumably as the ductus arteriosus closes. However, we are unable to quantify the f-h TD accurately at this time. Since the Pi-Top™ can only receive one Bluetooth data package at a time, there is a Bluetooth transmission difference between the hand and foot waveforms that will need to be accounted for in future analyses. Additionally, as a baby’s physiology changes and the potential for physiological foot peaks occurring before the hand peaks, this further complicates the analysis. However, the photoplethysmography segments shown in Fig. 4A and B were chosen because the relative hand to foot peak durations were consistent for these measurements. Thus, assuming a similar Bluetooth transmission across patients, this highlights the potential for a longer f-h TD in patients with CoA. We also captured a morphology change to the hand photoplethysmography waveform as the ductus arteriosus closed with the development of a remarkably notable dicrotic notch (Fig. 4C). This highlighted the need to apply smoothing filters to the waveforms so that peaks can more easily be identified to ease analysis.
Challenges and Adjustments Made
We encountered a few challenges, from which others will benefit from our experience. First, in regards to data security, we needed an encrypted USB compatible with Linux operating system and functional on an ARM processor (Raspberry Pi). This required using VeraCrypt™ encryption service as opposed to an off-the-shelf USB.
Additionally, to ensure the cessation of the Bluetooth connection, the entire Pi-top™ system has to be shut down. If the Bluetooth connection remains, then the data collection can continue when not intended. This is only problematic if data collection has to be restarted. For example, if a technical error is encountered mid-data collection, then the system has to be powered down and restarted, which adds a couple of minutes to the process.
To better identify collected data with specific clinical scenarios, such as the age of the patient at time of the measurement, we added an alphanumeric entry that serves as a code specific to the protocol time points. We were unable to use the time-stamps on the collected data, as some sites requested we falsify the date and time on the Pi-top™ to add additional security and completely de-identify the data. Even if the time-stamps were not falsified, the time-stamps alone would not be sufficient for identification due to time drift caused by not having the Pi-top™ connected to a server for security purposes [Reference Mills16].
Discussion
We created a system for automated collection of pulse oximetry data for research related to CCHD screening. This system collects and displays real-time data from medical devices while communicating wirelessly via Bluetooth. The system does not require access to a wireless network making it portable to the regions with little or no access to the internet and also aids with security. It is inexpensive and capable of end-to-end automation of data collection and storage. The system requires only one clinician or coordinator with basic computer skills to monitor the process. Once it is set up properly, the system can capture oxygen saturation and perfusion data from the infant in a non-invasive manner, and is able to maintain continuous operation for as long as required by the study circumstances.
Our data collection system is motivated by the need for early CCHD detection, and the inability for providers to electronically store pulse oximetry data in a manner conducive to the research and clinical needs. Antenatal echocardiology and postnatal examination detected only approximately 70% of the patients with CCHD leading to the addition of postnatal pulse oximetry to routine newborn screening [Reference Ailes, Gilboa, Honein and Oster6, Reference Sebelius17]. Techniques described in this study address the limitations of the oxygen saturation-based CCHD screen. Researchers have hypothesized that adding non-invasive pulse oximetry measurements such as PIx, radiofemoral pulse delay (f-h TD), and other photoplethysmography waveform characteristics to the current diagnostic suite of pulse oximetry measurement will help in more accurate detection of CCHD in patients [Reference Siefkes10–Reference Sorensen, Sadiq, Clifford, Maher and Oster14]. In a study with a total of 123 normal and 21 newborns with congenital heart defects, 4 out of 5 critical systemic obstruction subjects passed the regular oxygen saturation-based CCHD screen [Reference Siefkes10].The addition of PIx to the screen resulted in four of the five newborns failing the combined screen [Reference Siefkes10]. That study and prior studies evaluated PIx for a single moment in time with an artifact-free waveform for 10 s. In our current study, we averaged the pulse amplitude index (PAI), which is PIx multiplied by 100, over 5 min. We captured a patient with CoA and demonstrated the foot PAI decrease, as the ductus arteriosus was closing, and decreased below thresholds that may be indicative of systemic obstruction such as CoA [Reference de-Wahl Granelli and Ostman-Smith11, Reference Siefkes18]. Interestingly, we noted this decrease in PAI when the echocardiogram estimated only a mild CoA and thus it was initially thought the baby may not require surgery. Therefore, additional non-invasive perfusion measurements along with the regular oxygen saturation-based screen is a potential technique to not only enhance CCHD detection, but may also help guide treatment plans when it is already known that a patient has a CoA, although more research is necessary. Additionally, prior studies have noted prolonged pulse delay and abnormalities in timing of pre-ductal dichroitic notch in newborns with CoA [Reference Palmeri13, Reference Oyonarte, Dickinson, Medici and Hamilton19]. Our data captured an identifiable pre-ductal dicrotic notch while an infant with CoA was trialed off prostaglandin therapy and the timing relationship between the hand and foot pulses also appeared to change as well. However, conclusions cannot be drawn yet on this small sample of patients and additional analyses to sort out Bluetooth transmission versus physiological factors for the pulse delay are underway.
Future research into CCHD screening techniques will necessitate readily accessible medical device data, data standards, and archival data storage of identifiable data (identifiable at least through unique identification codes) [Reference Oster20]. The use of pulse oximetry devices results in huge amounts of data that are yet to be fully explored for CCHD screening. The current infrastructure limits access to this data for research. For example, continuous pulse oximetry monitoring in the hospital may not be stored long term or be readily identifiable, and when it is, researchers have to rely on charting to correlate with clinically relevant data such as probe placement [Reference Sorensen, Sadiq, Clifford, Maher and Oster14]. Our pulse oximeter data collection process mitigates many of these barriers and may also help guide future screenings or research for other diseases that may benefit from non-invasive pulse oximetry perfusion diagnostics (aneurysms, aortic dissections, atherosclerosis, thrombosis, or vascular graft monitoring).
Our system, while providing solutions to the above problems, is currently limited to the collection of pulse oximetry and perfusion information from a single type of medical device (Nonin® WristOx2™ 3150). Utilizing other devices, and other communication protocols besides Bluetooth is currently unsupported, and will require additional development efforts. Currently, CCHD screening capabilities are not offered by this system, but will be part of our future work. It is only designed to collect data from neonates who have already undergone or will undergo standard of care CCHD screening by a qualified provider to generate labeled datasets for research purposes, including the development of advanced machine learning-based CCHD detection models. Furthermore, no analytic capabilities for individual data streams such as perfusion are built into the system and our data visualization only displays streaming waveform data to the provider. Future work will serve to utilize this streaming waveform data in an analytic capacity and possibly even perform CCHD screening interpretation. However, additional studies and clinical trials will be required to prove its efficacy.
Overall, the system that we created has opened up a new way of data collection for CCHD related diagnostic information from neonates. The data collection system and process are mostly automated and are capable of being used by non-technical users. It may also serve as a generalized model to be utilized by different researchers who are also working to collect streaming waveform information. Using this system we have gathered approximately 1665 min of oximetry data from neonates with and without congenital heart disease. This information may be invaluable for developing future CCHD screening processes and hopefully will serve as the basis for future electronic systems to improve CCHD detection and may be adapted for other research endeavors as well.
Supplementary material
To view supplementary material for this article, please visit https://doi.org/10.1017/cts.2020.550.
Acknowledgments
We are grateful for the support of everyone who helped at various stages of this project and our study participants. Our engineers: Michael Agung, and research coordinators and collaborators: Michele Guillen, Orsolya Gilicze, Sparsha Govardhan, Alejandra Guzman, Erlinda Manalo, MD, Whitnee Hogan, MD, Robert Koppel, MD, and Meena Garg, MD.
The project described was supported by the National Center for Advancing Translational Sciences, National Institutes of Health (NIH), through grant number UL1 TR001860 and linked award KL2 TR001859, the Eunice Kennedy Shriver National Institute of Child Health & Human Development, NIH, through grant number 1R21HD099239–01, and the University of California, Davis Artificial Intelligence Seed Grant. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Disclosures
The authors do not have conflicts of interest to disclose. University of California, Davis has intellectual property rights related to inventions resulting from this work and a provisional patent has been submitted by the university.








