To the Editor—Clostridium difficile is an important cause of diarrhea in hospitals all over the world. It is among the most common organisms related to healthcare-associated infections in the United States and represents a serious public health concern.Reference Di Bella, Paglia, Johnson and Petrosillo 1 Clinical manifestations of C. difficile infection (CDI) range from asymptomatic carriage, mild or moderate diarrhea, to fulminant colitis.Reference Peng, Liu and Meng 2
In the last 20 years, hypervirulent isolates of C. difficile have been increasingly reported, mostly in North America and Europe.Reference Di Bella, Paglia, Johnson and Petrosillo 1 The main hypervirulent strain was named ribotype 027 (North American BI, NAP1/027). This epidemic strain was also reported in Asia, providing evidence of worldwide spread.Reference Aguayo, Flores and Lévesque 3 Patients infected with the 027 strain are 3 times more likely to have severe disease than those infected with non-027 strains.Reference Leffler and Lamont 4 , Reference Miller, Gravel and Mulvey 5 This finding was linked to increased production of A, B, and binary toxins, in association with a mutation in the gene regulating of the expression of these toxins, leading to overproduction. However, few studies have investigated the presence of hypervirulent strains of C. difficile in developing countries, particularly in Latin America.Reference Pires, Monteiro and Carneiro 6 , Reference Monteiro, Pires, Persson, Rodrigues Filho and Pasqualotto 7 Here, we make the first report of the detection of the hypervirulent C. difficile strain in Brazil.
METHODS
Setting and Patients
The hypervirulent C. difficile strain was noticed during the conduction of a point-prevalence multicenter study in Brazil. Fecal samples were tested with a commercial real-time PCR kit (Xpert C. difficile test; Cepheid, Sunnyvale, CA) in accordance with the manufacturer’s instructions. In addition, samples were also submitted to C. difficile culture, using absolute alcohol at room temperature, subcultured in CM0601 C. difficile agar (Oxoid), enriched with 7% blood horse, D-cycloserine, and cefoxitin for 48 hours under an anaerobic atmosphere. Species identification of suspected colonies was confirmed by matrix-assisted laser desorption/ionization (Brucker Daltonics, Bremen, Germany).
Presumptive diagnosis of infection with the 027 strain was conducted with GeneXpert and confirmed with in-house polymerase chain reaction (PCR), designed to amplify the tcdA and tcdB toxin genes and tcdC negative regulator toxin gene.Reference Griffiths, Fawley and Kachrimanidou 10 The PCR products were purified using the enzyme Exo-SAP-IT (Thermo Fischer, Waltham, MA) and sequenced using the BigDye Terminator version 3.1 Cycle Sequencing Kit (Life Technology, Carlsbad, CA). Clostridium difficile ATCC 9689 was used as a control. Sequence type was determined by multilocus sequencing typing,Reference Griffiths, Fawley and Kachrimanidou 10 and an online database was used to assign sequence typing (http://pubmlst.org/cdifficile).
RESULTS
The index case with a GeneXpert 027 positive result was recovered from a 68-year-old man who had a 2-month history of diarrhea and was heavily exposed to antimicrobial drugs (ie, vancomycin, metronidazole, amikacin and meropenem). He was admitted to a university hospital in Porto Alegre, Southern Brazil. At the time of sample collection, the patient had a C-reactive protein level of 109.7 mg/L and a leukocyte count of 8,000 cells/mL. The isolate harbored tcdA and tcdB toxin genes and the tcdC deletion (frameshift deletion, 18 nucleotides) that indicates toxin overproduction (Figure 1). The isolate was classified as ST67, belonging to clade 2. This clade contains a high diversity of sequence typing that also includes ST1 (NAP1/027).
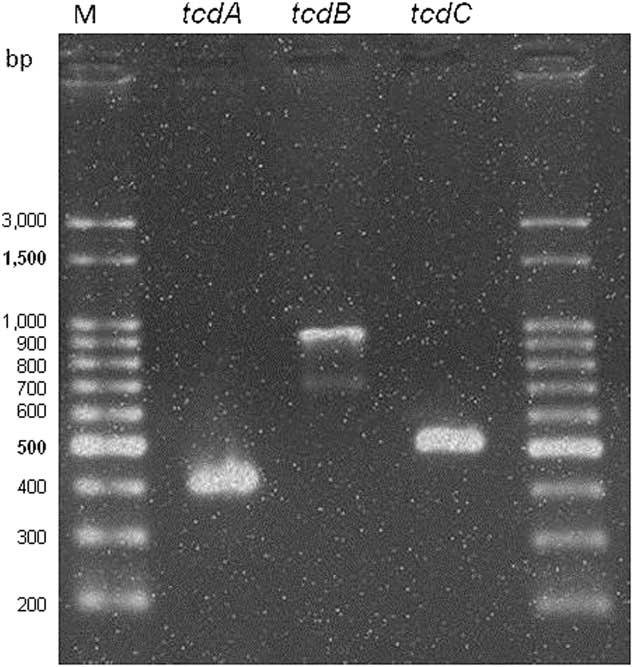
FIGURE 1 In-house polymerase chain reaction (PCR) test results for toxin genes tcdA, tcdB, and the negative promoter toxin gene tcdC. Legend: M, 100-bp size DNA ladder.
In another hospital located in the same city 1 month later, we identified another case of ribotype 027 using GeneXpert. Unfortunately, this case occurred in an outpatient, and we were unable to recover any sample for DNA sequencing. The patient had been using the following antibiotics >1 month for a difficult-to-treat otitis: amoxicillin, amoxicillin/clavulanate, and cefuroxime. She arrived in the emergency room with watery diarrhea lasting for 7 days. In the 3 days before admission, she reported abdominal pain, nausea, and malaise. Blood tests revealed 15,580 leucocytes/mL (81.4% neutrophils) and 349,000 platelets/mL. Abdominal ultrasound showed signs of colitis. The patient was discharged with metronidazole and was lost to follow-up.
DISCUSSION
A diagnosis of CDI is based on a combination of clinical history and laboratory detection of C. difficile toxins in the feces or cultured isolates. Nucleic acid amplification-based tests offer higher sensitivity and specificity. Although widely used for CDI diagnosis in developed countries, widespread implementation of such technologies in developing countries is still hampered by cost-related issues, in addition to the need of qualified infrastructure and staff.Reference Monteiro, Pires, Persson, Rodrigues Filho and Pasqualotto 7 In Brazil, immune enzymatic detection of C. difficile toxins remains the most widely used technique for CDI diagnosis because it is relatively inexpensive and easy to perform.
Diagnosis by GeneXpert offers a number of advantages, including agility, accuracy and presumptive detection of the hypervirulent strains with ~90% specificity.Reference Chiang, Ng, La, Jureen, Lin and Teo 8 On the other hand, a study reported unusual Xpert amplification curves from stool samples presumptively identified as 027/NAP1/BI but not confirmed by PCR ribotyping and tcdC gene sequencing.Reference Kok, Wang, Thomas and Gilbert 9 Also, in places with a low prevalence of hypervirulent strains, the positive predictive value of the test may be further reduced, reinforcing the importance of performing confirmatory tests. However, molecular typing of C. difficile isolates may not be available in all laboratories, and multiple strains of C. difficile may coexist in fecal samples.
The confirmed occurrence of hypervirulent C. difficile strain in Brazil emphasizes the need to improve surveillance strategies to potentially avoid outbreaks. Recently, Costa Rica, Chile, and Panama also reported isolation of the hypervirulent strain.Reference Aguayo, Flores and Lévesque 3 Ultimately, cost-effectiveness studies evaluating the use of real-time PCR to rapidly screen C. difficile infections are needed in Latin America. A recent study (69th ACC Annual Scientific Meeting, 2017, Abstract B-098) cited 10 cases of circulating 027 strains in the state of São Paulo, Brazil, but their finding was not confirmed by DNA sequencing and no epidemiological data were provided. Our data highlight the importance of testing for hypervirulence strains of C. difficile in patients diagnosed with CDI.
ACKNOWLEDGMENTS
Financial support: No financial support was provided relevant to this article.
Potential conflicts of interest: All authors report no conflicts of interest relevant to this article.





