1. Introduction
The discovery of RNA interference (RNAi) has opened a new perspective in genetics. This important mechanism modulates the expression of genes, helps cells defend themselves against viruses and regulates the activities of transposable elements. Some evidence indicates that RNAi also affects chromatin structure (Pal-Bhadra et al., Reference Pal-Bhadra, Leibovitch, Gandhi, Rao, Bhadra, Birchler and Elgin2004; Verdel et al., Reference Verdel, Jia, Gerber, Sugiyama, Gygi, Grewal and Moazed2004). In Drosophila melanogaster, for example, mutations in genes that control RNAi may alter the distribution of chromatin-organizing proteins such as HP1 and histone acetyltransferase, even when the mutations are heterozygous. In addition, some of these mutations cause the variegated expression of certain transgenes to be more like wild-type – a phenomenon thought to involve changes in chromatin structure (Pal-Bhadra et al., Reference Pal-Bhadra, Leibovitch, Gandhi, Rao, Bhadra, Birchler and Elgin2004). In these respects, RNAi mutations resemble the suppressor of variegation, or Su(var), mutations, some of which also appear to alter the frequency and distribution of crossing over during meiosis (Westphal & Reuter, Reference Westphal and Reuter2002). These observations raise the possibility that RNAi mutations might likewise alter the pattern of crossing over – an outcome that would imply a role for RNAi in the structure and/or behaviour of chromosomes in germ-line cells. To investigate this possibility, we have tested a sample of RNAi mutations for altered patterns of crossing over in the euchromatin of the X chromosome and in the centric heterochromatin of chromosome II.
The mutations tested involve three genes: aubergine (aub), piwi and homeless (hls; also known as spindle-E). The genes aub and piwi encode Argonaute-type proteins, which are known to be involved in RNAi pathways in several different organisms (Brennecke et al., Reference Brennecke, Aravin, Start, Dus, Kellis, Sachidanandam and Hannon2007). These proteins bind small interfering RNAs and guide them to their larger mRNA targets, which may then be destroyed. The hls gene encodes an RNA helicase that also appears to be involved in RNAi (Usakin et al., Reference Usakin, Abad, Vagin, de Pablos, Villasante and Gvozdev2007). In Drosophila, the Aub, Piwi and Hls proteins are produced in the female germ line (Brennecke et al., Reference Brennecke, Aravin, Start, Dus, Kellis, Sachidanandam and Hannon2007; Usakin et al., Reference Usakin, Abad, Vagin, de Pablos, Villasante and Gvozdev2007), where they seem to play an important role in transposon regulation (Vagin et al., Reference Vagin, Sigova, Li, Seitz, Gvozdev and Zamore2006). Their presence in the female germ line supports the idea that RNAi might influence meiotic crossing over, which in D. melanogaster occurs only in females.
Because RNAi mutations are either lethal or female sterile in homozygotes, they cannot be tested for effects on recombination in double dose. However, a set of these mutations has previously been assayed for impairment of transposon regulation, and two of them, aub ΔP-3a and aub QC42, were found to disrupt this regulation through dominant effects (Simmons et al., Reference Simmons, Ryzek, Lamour, Goodman, Kummer and Merriman2007). This finding, along with other evidence for dominant effects (Pal-Bhadra et al., Reference Pal-Bhadra, Leibovitch, Gandhi, Rao, Bhadra, Birchler and Elgin2004), raised the possibility that mutations in RNAi genes might, like some Su(var) mutations, influence the pattern of crossing even when they are heterozygous. Thus, our experimental strategy was to cross females from the mutant stocks to males with genetic markers on the X or second chromosome and then backcross their daughters, which were heterozygous for the maternally inherited RNAi mutation and the markers on the X or chromosome II, to marked males. The offspring of these backcrosses were scored for recombination in the chromosomal regions delimited by the markers.
2. Materials and methods
2.1. Drosophila stocks and husbandry
The genetic markers and special chromosomes used in the experiments are fully described in the Flybase website, in Lindsley & Zimm (Reference Lindsley and Zimm1992) and in other references cited in the text. The X-linked markers (and their standard map positions) were y (0·0), w (1·5), sn w, sn 3 (21·0), v (33·0) and car (62·5). These markers span most of the euchromatic portion of XL. The II-linked markers were lt and rl. These markers lie at opposite ends of the centric heterochromatin, which constitutes about one-third of the chromosome's overall length; however, because of the rarity of crossing over in this region, lt and rl are very tightly linked to each other. Experimental cultures were reared in shell vials at 25°C on a standard cornmeal, molasses, dried yeast and agar medium. Each test culture was established with a single female and 2–3 males, and after 1 week, the parents were transferred to a fresh culture vial. The progeny of the original and transfer test cultures were scored for recombinants 2 weeks after they were started.
2.2. Experimental procedures
Mutations in aub, hls and piwi were tested for effects on crossing over in the euchromatin of the long arm of the X chromosome, and mutations in aub and piwi were tested for effects on crossing over in the centric heterochromatin of chromosome II.
To study crossing over in the X chromosome, test females were obtained from mass matings between either (1) w sn w or (2) w sn w; RNAi mutation/balancer females and y sn 3v car males. Cross (1) provided +w+sn w++/y+sn 3v car females that were used as controls in the study. Cross (2) provided +w+sn w++/y+sn 3v car females that were heterozygous for an autosomal RNAi mutation or the balancer chromosome used to maintain that mutation in stock. For the aub and piwi mutations, the balancer was Cy Roi [=In(2L)Cy Lt R+In(2R)Cy, Cy Roi cn sp bw], and for the hls mutations it was TM6, Tb. Because these balancers are multiply inverted, they might be expected to increase the frequency of crossing over in other chromosomes through an interchromosomal effect (Lucchesi & Suzuki, Reference Lucchesi and Suzuki1968). Females heterozygous for the balancer chromosomes therefore served as positive controls in the study. All the +w+sn w++/y+sn 3v car females produced by crosses (1) and (2) were mated to y sn 3v car males and the offspring were scored for recombination among the X-linked markers. Because of epistasis of the w marker over the v and car markers, sons were scored for crossovers to the left of sn and daughters were scored for crossovers to the right of sn.
A follow-up experiment was carried out to determine if one of the balancer chromosomes could influence crossing over in the X chromosome through a maternal effect. w sn w; +/Cy Roi females from a cross between w sn w; Gla/Cy Roi females and w sn w males were mated to y sn 3v car males and their two kinds of daughters, +w+sn w++/y+sn 3v car; +/(+) and +w+sn w++/y+sn 3v car; +/Cy Roi, were backcrossed to y sn 3v car males to obtain the flies that were scored for recombinants. The (+) in the genotype of the first kind of daughter denotes the +chromosome inherited from the +/Cy Roi mother; even though this class of daughters does not carry the Cy Roi balancer chromosome, it could have an altered frequency of crossing over in the X chromosome if the balancer exerted a maternal effect. The controls in this experiment were +w+sn w++/y+sn 3v car females that came from a cross between w sn w females and y sn 3v car males.
To study crossing over in the centric heterochromatin of chromosome II, females from an RNAi mutation/CyO stock were crossed to b lt rl males, and then the RNAi mutation/b lt rl daughters were backcrossed to b lt rl males to obtain progeny that were scored for recombination between lt and rl. As controls, we tested +/b lt rl females that were obtained from a two-generation scheme; aub QC42/CyO females were crossed to males from the wild-type strain Samarkand and their +/CyO daughters were crossed to b lt rl males to obtain the females for testing. We also tested +/b lt rl females that were heterozygous for TM3, Sb, a balancer chromosome III, to determine if recombination between lt and rl is influenced by an interchromosomal effect; these test females were obtained by crossing hls E616/TM3, Sb females to b lt rl males.
Statistical significance was assessed by performing z-tests using standard errors calculated from binomial and large-sample variances.
3. Results
3.1. Effects on recombination in the euchromatin of the X chromosome
Mutations in the aub, hls and piwi genes and the balancer chromosomes used to maintain them were tested for possible effects on the pattern of crossing over in the euchromatic portion of XL. It should be noted that the tested females in all the experimental groups were heterozygous for the same two X chromosomes, w sn w and y sn 3v car. The results of all these tests are summarized in Table 1.
Table 1. Effects of RNAi mutations and balancer chromosomes on recombination in the euchromatic X chromosome

a Each map distance, in centiMorgans, was estimated by the observed frequency of recombination in the interval. The standard error was computed from the binomial variance of the recombination frequency. For intervals longer than about 20 cM, map distances are expected to be underestimated due to the occurrence of undetected double exchanges within them.
b The total map distance is in centiMorgans. The standard error was computed from the sum of the binomial variances of the constitutive recombination frequencies.
c These groups come from an experiment to test for a maternal effect of the Cy Roi balancer chromosome on recombination in the X chromosome. Females in the group that might show the maternal effect, denoted (+), inherited a wild-type chromosome II from their +/Cy Roi mothers.
The females without any mutation or balancer chromosome in their genotype – i.e. the wild-type controls – gave the shortest total map length, 50·79 cM, and females with a balancer chromosome in their genotype gave the longest, 65·60 cM for Cy Roi and 65·30 cM for TM6, Tb. The maps from the flies with the balancer chromosomes are significantly longer than those from any other genotype except piwi 2. Thus, each of the balancers increases the frequency of recombination in the X chromosome – a clear example of the classical interchromosomal effect. This greater frequency of recombination in the presence of a balancer chromosome reflects a significantly higher incidence of crossing over in three of the four intervals on the X: y-w, w-sn and v-car. Only the centrally located sn-v interval did not seem to undergo more crossing over when a balancer chromosome was present in the genotype.
Females heterozygous for the RNAi mutations gave intermediate map lengths, ranging from 52·16 for hls E616 to 59·67 for piwi 2. The maps for the aub QC42, piwi 1 and piwi 2 mutations are significantly longer than the map of the wild-type controls. For the piwi 2 mutation, the map intervals that showed increased recombination were the same as the ones that showed it for the balancer chromosomes, although only two of them (y-w and w-sn) showed statistically significant increases. For the aub QC42 mutation, only the w-sn interval showed a significant increase in recombination frequency. In the case of the piwi 1 mutation, none of the individual map intervals showed a statistically significant increase, but the cumulative effect of the increases in each of them made the total map length significantly greater than that of the wild-type controls.
These data suggest that at least some of the RNAi mutations increased crossing over in the X chromosome. However, it is possible that the observed increases in total map length are due to maternal effects of the balancer chromosomes, which were present in the mutant stocks, rather than to effects of the RNAi mutations themselves. To test this possibility, we assessed recombination in the X chromosome in the two types of daughters produced by +/Cy Roi females: those that inherited the Cy Roi balancer chromosome, which would be expected to show increased recombination in the X chromosome due to an interchromosomal effect, and those that inherited the +chromosome, which would only show increased recombination in the X if the Cy Roi balancer exerted its recombination-enhancing effect maternally. The results of this experiment are summarized in the last three rows of Table 1.
The total map length from the wild-type controls (52·28 cM) was only slightly greater than that from the controls in the first experiment, and the map length from the Cy Roi flies (65·80 cM) was essentially the same as that from the Cy Roi flies in the first experiment. Thus, the genetic benchmarks from the first experiment (control map length and map length in the presence of a balancer chromosome) proved to be reproducible. To see if the balancer chromosome exerted a maternal effect on recombination in the X chromosome, we must focus on the map length from the wild-type sisters (denoted (+) in Table 1) of the Cy Roi females in the second experiment. This value (54·10 cM) lies between the map lengths of the wild-type controls and the Cy Roi flies, as might be expected if there is a maternal effect; however, 54·10 is not significantly different from 52·28, the map length of the wild-type controls. Thus, if the Cy Roi chromosome exerts a maternal effect, it is not nearly as strong as the chromosome's zygotic effect on recombination in the X. Note, however, that 54·10 cM is also not significantly different from the map lengths from the various mutant strains in the first experiment (52·16–59·67). Thus, using 54·10 cM as the basis for evaluating these map lengths – and this procedure takes into account the possibility of a maternal effect of the balancer chromosome – there is not strong evidence that any of the RNAi mutations increased recombination in the X chromosome in the first experiment.
The data from the two recombination experiments were also used to examine the coincidence of crossovers in the sn-v and v-car intervals on the X chromosome (Table 2). Compared with the wild-type controls, the flies with balancer chromosomes in their genotypes had markedly greater frequencies of double crossovers in these intervals – another manifestation of the recombination-enhancing effect of a balancer on one of the autosomes. The coefficients of coincidence for the flies with the balancers were also inflated, more so in the second experiment than in the first. Crossing over in the X chromosome therefore appears to be freer – i.e. less subject to interference – when an interchromosomal effect is in play. The data from the (+) flies in the second experiment also suggest that both of these measures of recombination could have been influenced by a maternal effect of the balancer chromosome, although the differences between the+and (+) flies and between the (+) and Cy Roi flies are not statistically significant. Furthermore, when we compare the frequencies of double crossovers or the coefficients of coincidence from the flies with the RNAi mutations with those from the wild-type controls in the first experiment, there are no significant differences. Thus, the RNAi mutations apparently do not influence the frequency of double crossing over or the amount of interference in the X chromosome.
Table 2. Frequency of double crossovers in the interval between sn and car on the X chromosome

a The coefficient of coincidence (c) was computed as the ratio of the observed frequency of double crossovers (z) to the expected frequency of double crossovers. This latter frequency was calculated from the recombination frequencies between sn and v (x) and between v and car (y) under the assumption that the crossovers in these two intervals occur independently, i.e. with frequency x×y. Thus, c=z/(x×y). The standard error of c was obtained from its large-sample variance, estimated as V(c)=c 2[CV2(x)+CV2(y)+CV2(z)], where the CVs are the coefficients of variation of the constitutive statistics.
b These data were obtained from the experiment to test for a maternal effect of the Cy Roi balancer chromosome.
3.2. Effects on recombination in the centric heterochromatin of chromosome II
The forgoing analyses attempted to assess if RNAi mutations influence the pattern of recombination in the euchromatin, where crossing over typically occurs. We also tested for an influence in the centric heterochromatin, where crossing over is exceedingly rare. The results of these tests are summarized in Table 3.
Table 3. Effects of aub and piwi mutations on crossing over in the centric heterochromatin of chromosome II
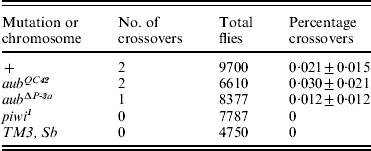
Only three mutations were examined in this experiment: aub QC42, aub ΔP-3a and piwi 1. None of them seemed to have any effect on the frequency of crossing over between lt and rl, which was very low in all the test groups, and not significantly different from published estimates (Westphal & Reuter, Reference Westphal and Reuter2002). Thus, in these data there is no evidence that RNAi plays a role in the occurrence of crossing over in the centric heterochromatin, nor is there any evidence for a recombination-enhancing interchromosomal effect in this part of the genome.
4. Discussion
To the question posed by the title of the present paper, ‘Does RNA interference influence meiotic crossing over in Drosophila melanogaster?’, the answer appears to be no. We took a genetic approach to this question and found no strong evidence that crossing over is affected by mutations in key RNAi genes. The largest effect – a 9% increase in the map length of the euchromatic portion of the X chromosome – was seen in flies that carried the piwi 2 mutation; however, this effect was not statistically significant, either because of intrinsic variation in the frequency of recombination in the X or because of a maternal interchromosomal effect of the balancer chromosome that was used to maintain the piwi 2 mutation in stock.
The conclusion that RNAi does not influence crossing over must, however, be taken cautiously. One important caveat is that the various RNAi mutations could only be studied under heterozygous condition. Under homozygous condition, they are either lethal or female sterilizing. Thus, it was not possible to knock out the RNAi machinery; the best we could do was to knock it down by mutating one copy of an RNAi gene. However, such knock-down experiments have demonstrated that RNAi is involved in the regulation of transposable elements (Reiss et al., Reference Reiss, Josse, Anxolabéhère and Ronsseray2004). Mutations in aub, for example, completely disrupt cytotype regulation of P elements through dominant, maternal effects (Simmons et al., Reference Simmons, Ryzek, Lamour, Goodman, Kummer and Merriman2007). Thus, the approach taken here – to look for dominant effects of the RNAi mutations on crossing over – is founded on a solid precedent. Another caveat is that the pattern of crossing over could be controlled by redundant RNAi pathways, or by redundant components of one pathway, such that a mutation in a single gene would not produce a noticeable effect. The epistasis implied by this scenario could be investigated by testing two or more mutations simultaneously. A third caveat is that only three RNAi genes were studied. Other RNAi genes have been identified, and mutations are available for some of them. These mutations could be tested for effects on crossing over in the future. However, the three genes analysed in the present study were arguably the best candidates among all the RNAi genes known, and the negative results obtained with them suggest that RNAi simply does not influence crossing over.
RNAi may, however, still have some effect on chromosome structure, and if not in the germ line, then in somatic cells, where there is evidence that it affects the distribution of certain proteins along the chromosomes (Pal-Bhadra et al., Reference Pal-Bhadra, Leibovitch, Gandhi, Rao, Bhadra, Birchler and Elgin2004). Large RNAs clearly can affect the appearance and functionality of chromosomes. For example, the diffuse appearance of the X chromosome in male Drosophila and the hyperactivation of its genes are caused by the roX RNAs in co-operation with a small set of proteins (Kelley & Kuroda, Reference Kelley and Kuroda2000), and the condensation of one of the X chromosomes in female mammals and the inactivation of most of its genes are initiated by the XIST RNA transcribed from the X destined to be inactivated (Panning & Jaenisch, Reference Panning and Jaenisch1998). If these large RNAs can influence chromosome structure, then perhaps small interfering RNAs can do likewise.
Although we did not find evidence that RNAi mutations affect crossing over, we did show that balancer chromosomes affect it, specifically that crossing over in the euchromatic portion of the X chromosome is enhanced by a balancer chromosome elsewhere in the genome – the classic interchromosomal effect. This enhancement of crossing over was seen in the distal and proximal regions of the X chromosome, but not in the central region between the markers sn and v. Portin & Rantanen (Reference Portin and Rantanen1990) observed a similar pattern. We also found less interference in the X chromosome (between sn and car) when an autosomal balancer was present, as did Portin & Rantanen (Reference Portin and Rantanen1990), although they studied a different region in the X chromosome (between cv and f). In another analysis, Sniegowski et al. (Reference Sniegowski, Pringle and Hughes1994) found that autosomal inversions increased recombination at the base of the X chromosome (between f and su(f)), but not at the tip (between y and spl). We did not test specifically for a recombination-enhancing interchromosomal effect at the base of the X chromosome – the interval from v to car that we tested is much broader, but we did find an effect at the tip, although only in the first experiment. In the second experiment, the Cy Roi balancer actually seemed to decrease recombination there. We also did not see increased recombination in the centric heterochromatin of chromosome II when a balancer chromosome III was present in the genotype. However, other investigators have seen increased recombination in this region in the presence of a rearrangement elsewhere (reviewed in Lucchesi & Suzuki, Reference Lucchesi and Suzuki1968).
We conducted an experiment to determine if the interchromosomal effect is transmitted maternally to offspring that do not themselves inherit a rearranged chromosome. This maternal effect could explain the apparent increases in recombination that we saw in females whose mothers were heterozygous for an RNAi mutation and a balancer chromosome, and who inherited the RNAi mutation rather than the balancer chromosome. Such a maternal effect might also explain the increase in recombination that has been attributed to some Su(var) mutations (Westphal & Reuter, Reference Westphal and Reuter2002). The results of this experiment were consistent with there being a weak maternal interchromosomal effect, which, however, was not statistically significant.
A maternal component of the interchromosomal effect would, of course, rule out explanations for recombination enhancement based on unusual chromosomal associations caused by a rearranged chromosome in the genome (reviewed in Lucchesi & Suzuki, Reference Lucchesi and Suzuki1968). An alternative explanation is that the rearranged chromosome causes a redistribution of chromosomal proteins throughout the genome, perhaps because the rearranged chromosome has an abnormal pattern of euchromatin/heterochromatin junctions. Proteins such as HP1, which plays a key role in heterochromatin formation (James et al., Reference James, Eissenberg, Craig, Diethrich, Hobson and Elgin1989; Eissenberg et al., Reference Eissenberg, James, Foster-Hartnett, Hartnett, Ngan and Elgin1990; Danzer & Wallrath, Reference Danzer and Wallrath2004), could become more concentrated on the rearranged chromosome by spreading from heterochromatin into euchromatin at all the junctions, and therefore become less concentrated on other chromosomes. If this altered – i.e. looser – packaging on the other chromosomes persists, it could lead to increased crossing over during meiosis in individuals of the next generation, even when they have not inherited the rearranged chromosome. Thus, the interchromosomal effect might be mediated by epigenetic changes in chromosome packaging induced by a rearranged chromosome somewhere in the genome.
James Birchler and Barbara Wakimoto kindly provided Drosophila stocks. E. W. C. was supported by the Undergraduate Research Opportunities Program of the University of Minnesota. Further support came from the Department of Genetics, Cell Biology and Development and the College of Biological Sciences of the University of Minnesota.







