1. Introduction
Chromosomes are understood to be positioned in particular territories within the nucleus (Manuelidis, Reference Manuelidis1985; Schardin et al., Reference Schardin, Cremer, Hager and Lang1985). These help maintain efficient gene expression (Kosak & Groudine, Reference Kosak and Groudine2004) and may therefore be involved in gene regulation (Fraser & Bickmore, Reference Fraser and Bickmore2007; Noordermeer et al., Reference Noordermeer, Branco, Splinter, Klous, van IJcken, Swagemakers, Koutsourakis, van der Spek, Pombo and de Laat2008). A role for chromosomal territories (CTs) in determining the likelihood of translocation of particular chromosomes has been suggested (Branco & Pombo, Reference Branco and Pombo2006), and demonstrated, in the case of non-Robertsonian (Rb) translocation in humans (Ashley et al., Reference Ashley, Gaeth, Inagaki, Seftel, Cohen, Anderson, Kurahashi and Emanuel2006). Importantly, however, the arrangement of chromosomes varies not only by cell type but also by cell cycle phase. Although CTs have mostly been studied in interphase and mitotic prometaphase of somatic cells, the germ cell path (the only one relevant to the persistence of Rb fusions, i.e. whole arm fusions) has recently received increasing attention (e.g. Garagna et al., Reference Garagna, Zuccotti, Thornhill, Fernandez-Donoso, Berrios, Capanna and Redi2001; Merico et al., Reference Merico, Pigozzi, Esposito, Merani and Garagna2003, Reference Merico, de Barboza, Vasco, Ponce, Rodriguez, Garagna and de Talamoni2008; Foster et al., Reference Foster, Abeydeera, Griffin and Bridger2005; Ashley et al., Reference Ashley, Gaeth, Inagaki, Seftel, Cohen, Anderson, Kurahashi and Emanuel2006; Berríos et al., Reference Berríos, Manterola, Prieto, López-Fenner, Page and Fernández-Donoso2010; Nunes et al., Reference Nunes, Catalan, Lopez, da Graça Ramalhinho, da Luz Mathias and Britton-Davidian2010; Vavouri & Lehner, Reference Vavouri and Lehner2011). This is important as chromosomes change their positioning during spermatogenesis (as evidenced in pigs; Foster et al., Reference Foster, Abeydeera, Griffin and Bridger2005) suggesting that Rb-relevant proximity relationships may be different during meiosis.
The exact location of territories can only be described probabilistically (Parada et al., Reference Parada, Roix and Misteli2003; Bolzer et al., Reference Bolzer, Kreth, Solovei, Koehler, Saracoglu, Fauth, Müller, Eils, Cremer, Speicher and Cremer2005). In fact, the proximity patterns among CTs have been described from the statistical analysis of populations of cells, rather than the consistent, repeatable behaviour of individual cells (Cremer & Cremer, Reference Cremer and Cremer2010). Furthermore, it has been observed that territories intermingle in interphase to the extent that in human lymphocytes an average of 46% of a chromosome is mixed in with other chromosomes. This corresponds to 2·1±1·1% (on average) for any given combination of chromosomes (Branco & Pombo, Reference Branco and Pombo2006). The significant intermingling of chromosomes has been reproduced in relatively coarse-grained cubic lattice-based Monte Carlo simulations that describe the decondensation at the beginning of interphase (Fritsch & Langowski, Reference Fritsch and Langowski2011). There has also been a report of a single territory that is shared by a cluster of chromosomes of similar size (Parada et al., Reference Parada, McQueen, Munson and Misteli2002). At the same time, homologous chromosomes can inhabit separate territories that may be some distance apart (Bolzer et al., Reference Bolzer, Kreth, Solovei, Koehler, Saracoglu, Fauth, Müller, Eils, Cremer, Speicher and Cremer2005) and small chromosomes, at least in a human lymphoblastoid cell line, had more physical contact with each other (Lieberman-Aiden et al., Reference Lieberman-Aiden, van Berkum, Williams, Imakaev, Ragoczy, Telling, Amit, Lajoie, Sabo, Dorschner, Sandstrom, Bernstein, Bender, Groudine, Gnirke, Stamatoyannopoulos, Mirny, Lander and Dekker2009). The exception to this is chromosome 18, which has the lowest gene density among the small human chromosomes. Large chromosomes also had slightly more contact with each other than expected by chance, and medium-sized chromosomes had less contact with other chromosomes as well as with each other (Lieberman-Aiden et al., Reference Lieberman-Aiden, van Berkum, Williams, Imakaev, Ragoczy, Telling, Amit, Lajoie, Sabo, Dorschner, Sandstrom, Bernstein, Bender, Groudine, Gnirke, Stamatoyannopoulos, Mirny, Lander and Dekker2009).
The area between CTs, the interchromatin domain (or interchromosome domain (ICD); Zirbel et al., Reference Zirbel, Mathieu, Kurz, Cremer and Lichter1993), is believed to have a special function. This is positioned directly between CTs and away from points of contact with the nuclear envelope and nucleolus (Scheuermann et al., Reference Scheuermann, Tajbakhsh, Kurz, Saracoglu, Eils and Lichter2004). The ICD is believed to be rich in molecular machinery that is essential for nucleic acid processing (Branco & Pombo, Reference Branco and Pombo2006 and references therein) and genes are mostly found at the periphery of a CT, while non-transcribed sequences (including X-inactivated genes; Dietzel et al., Reference Dietzel, Schiebel, Little, Edelmann, Rappold, Eils, Cremer and Cremer1999) localize more interiorly (Scheuermann et al., Reference Scheuermann, Tajbakhsh, Kurz, Saracoglu, Eils and Lichter2004). Active genes, however, concentrate in the ICD (Scheuermann et al., Reference Scheuermann, Tajbakhsh, Kurz, Saracoglu, Eils and Lichter2004).
Until recently it was thought that hybrids of different Rb chromosomal races would experience underdominance in the heterozygous state. Unexpectedly though, data from house mouse populations from the Island of Madeira suggest little such disadvantage, if any, while the two contacting populations differ only by the presence/absence of a single Rb fusion (Nunes et al., Reference Nunes, Catalan, Lopez, da Graça Ramalhinho, da Luz Mathias and Britton-Davidian2010). (However, see Merico et al., Reference Merico, Pigozzi, Esposito, Merani and Garagna2003, who show that male hybrids aged 3 and 5 months – rather than 7 months as used by Nunes et al. – have more seriously impaired germ cell survival). More severe reductions in fitness are seen when the same ancestrally acrocentric chromosome has undergone two different fusions in the contacting populations (i.e. monobrachial fusions). This carried a fitness penalty of one-third, suggesting selection for meiotic checks against Rb fusions. An increased incidence of particular Rb fusions could mean that these incur a lower penalty on fertility (or other aspects of fitness), or are created at higher rates, or some combination of the two.
Comprehensive Rb fusion datasets are available for mice and bovids, and these differ in several ways that should make this contrast interesting. Mice are small, shorter-lived, selected for high reproductive rates and have large effective population sizes, whereas bovids are generally much larger, longer-lived, of smaller population size, longer generation times and, relatively speaking, are selected for high survival rates (Romiguier et al., Reference Romiguier, Ranwez, Douzery and Galtier2010). The cattle genome is used here as it is currently the best studied in the group, and because it is believed to be somewhat conserved, reflecting, at least with respect to karyotype (Robinson & Ropiquet, Reference Robinson and Ropiquet2011 and references therein), the likely ancestral bovid condition.
The rationale for comparing the Rb dynamics in a set of mouse populations with those in a set of bovid species is that populations can form into species. In other words, if an aggregation of individuals is distinct enough to be recognized as a separate population, it can just as easily be thought of as a potential origin of a separate species-to-come. Hence, each of the datasets has unique advantages. The studied mouse populations give a cross-section through evolution over large geographic scales at a sampling density that far surpasses any bovid species, but with gene flow patterns that have not been fully elucidated. In contrast, the bovid phylogeny used here (51 extant, i.e. terminal node species) provides a large number of independent replicates of chromosomal evolution, as well as a larger interaction space with 29 ancestral autosomes, ten more than the house mouse.
Based on the hypothesis that chromosomes occupy territories whose position within the nucleus may correspond to chromosome size in a somewhat linear way (Bolzer et al., Reference Bolzer, Kreth, Solovei, Koehler, Saracoglu, Fauth, Müller, Eils, Cremer, Speicher and Cremer2005), we can formulate two competing hypotheses of what the shape of the function predicting Rb fusion participation frequency would be on the basis of chromosome size. The null hypothesis is that chromosomes encounter each other randomly as they move about the nucleus at the beginning of meiotic prophase I. Since the two chromosomes need to physically meet in order for fusion to occur, and larger chromosomes could present bigger targets, this null hypothesis would predict that large chromosomes are the most frequent Rb fusers, with medium-sized chromosomes intermediate and small chromosomes being rare to fuse.
Under the alternate, ‘physical proximity’ hypothesis, particular spatial relationships such as CTs would be enforced in early prophase. As a result, both large and small chromosomes would be in marginal positions, and medium-sized chromosomes would be in a position that puts them between the other two groups. Hence, medium-sized chromosomes would have opportunities to fuse with both of the other groups, whereas large chromosomes would be prevented from meeting small chromosomes (and vice versa), and so both groups would have smaller Rb fusion frequencies.
In this paper, we use data from the mouse and bovids to investigate how nuclear positioning of chromosomes affects the frequency of participation of autosomes in Rb fusions. We focus, in particular, on chromosome size and GC content as previously proposed determinants of chromosomal positioning in the nucleus and find that size is a better predictor of Rb fusions in bovids, at the elimination of GC content. We then point out differences in chromosome participation and partnering patterns between mouse and bovids, and review the findings in terms of previous evidence of chromosomal positioning.
2. Methods
(i) Genomic data
We used the bovine genome assembly version Btau_4.0 that is available in GenBank (accession number AAFC0000000.3). GC content was computed from a sequence that was not repeat-masked using a simple Perl script. Chromosome lengths were calculated using Unix shell tools. Mouse genomic data, where mentioned, were taken from NCBI build 37. For mouse housekeeping (HKP) genes, the more stringent data from Ruiz-Herrera et al. (Reference Ruiz-Herrera, Farré, Ponsà and Robinson2010) were re-used, while gene density for both species was calculated from data accessible through the ENSEMBL genome browser, using only genes of known function (supplementary Table S1 available online at http://journals.cambridge.org/grh). Briefly, Ruiz-Herrera et al. (Reference Ruiz-Herrera, Farré, Ponsà and Robinson2010) classified HKP genes as those that were of known function and had expression levels above the median in all of the 61 tissues included in the Gene Expression Atlas (Su et al., Reference Su, Wiltshire, Batalov, Lapp, Ching, Block, Zhang, Soden, Hayakawa, Kreiman, Cooke, Walker and Hogenesch2004).
(ii) Positional data
In this study, we follow Mayer et al. (Reference Mayer, Brero, von Hase, Schroeder, Cremer and Dietzel2005) who determined the position of five autosomes (which form the focus here) and the X chromosome in four different tissue types and cell cycle stages of the house mouse. Other tissue type/phase combinations had insufficient sample sizes. Mayer et al. (Reference Mayer, Brero, von Hase, Schroeder, Cremer and Dietzel2005) restricted their observations to round nuclei for technical reasons. These authors present several median values for each combination of chromosome and cell type/cell cycle phase combination, whereof we use the mean values.
(iii) Chromosomal data
Chromosomal characters for 51 bovids were taken from Robinson & Ropiquet (Reference Robinson and Ropiquet2011), and those for the mouse from Piálek et al. (Reference Piálek, Hauffe and Searle2005) . Since Rb fusion frequency is potentially confounded by the opportunities a particular chromosome has to fuse based on the branching patterns within a phylogenetic tree, we computed these using a bovid consensus phylogeny (see Figure 2 in Robinson & Ropiquet Reference Robinson and Ropiquet2011 and source references therein) as the evolutionary backbone against which the respective fusions were mapped. Piálek et al. (Reference Piálek, Hauffe and Searle2005) compiled data on occurrence of Rb fusions in 97 surveyed mouse populations. We follow Gazave et al. (Reference Gazave, Catalan, da Graça Ramalhinho, da Luz Mathias, Nunes, Dumas, Britton-Davidian and Affray2003) in analysing the mouse data in terms of whether a fusion has been observed, as well as how many populations were observed to be carrying it. Assuming independence of mouse populations, we worked with a set of 479 individual fusion events in the house mouse and 195 in bovids (99 and 141 unique changes, respectively; Table 1).
Table 1. Summary of Rb fusion datasets (raw data for bovids described in Robinson and Ropiquet (Reference Robinson and Ropiquet2011) and house mouse in Piálek et al. (Reference Piálek, Hauffe and Searle2005) )

Other events that affected the availability of acrocentric autosomes (i.e. non-Rb fusions and fusions of autosomes with sex chromosomes) were also considered; all other characters were ignored. The ratios of participations over opportunities were computed and log-transformed for further analysis. Summary data on the bovid and mouse datasets are given in Table 1.
(iv) General linear model (GLM) statistics
We used linear regression complemented with coefficients of correlation (after Pearson unless stated otherwise), as well as GLMs to test variables previously hypothesized (or observed) to explain either Rb fusion frequency or chromosomal positioning as predictors of Rb fusion frequency. In bovids, we were also able to control for the opportunities to fuse as dictated by the shape of the phylogeny (see above). Our aim was to explain the number of Rb fusion events per chromosome as a function of other factors. Previous studies found cell types to differ in whether chromosomes are arranged in a radial pattern according to gene density (Boyle et al., Reference Boyle, Gilchrist, Bridger, Mahy, Ellis and Bickmore2001; Gilbert et al., Reference Gilbert, Gilchrist and Bickmore2005; Küpper et al., Reference Küpper, Kölbl, Biener, Dittrich, von Hase, Thormeyer, Fiegler, Carter, Speicher, Cremer and Cremer2007) of which GC content is a close correlate (e.g. Mouse Genome Sequencing Consortium, 2002; Lercher et al., Reference Lercher, Urrutia and Hurst2002), or chromosome size (Sun et al., Reference Sun, Shen and Yokota2000; Cremer et al., Reference Cremer, von Hase, Volm, Brero, Kreth, Walter, Fischer, Solovei, Cremer and Cremer2001; Bolzer et al., Reference Bolzer, Kreth, Solovei, Koehler, Saracoglu, Fauth, Müller, Eils, Cremer, Speicher and Cremer2005). It has been suggested that chromosome size is a stronger predictor in flat-ellipsoidal nuclei, whereas GC content and gene density have greater influence in spherical nuclei (Bolzer et al., Reference Bolzer, Kreth, Solovei, Koehler, Saracoglu, Fauth, Müller, Eils, Cremer, Speicher and Cremer2005; Skinner et al., Reference Skinner, Völker, Ellis and Griffin2009), but both are significant predictors across a range of cell types and shapes (Mayer et al., Reference Mayer, Brero, von Hase, Schroeder, Cremer and Dietzel2005). Therefore, for bovids, the dependent variables were (a) the number of fusion events by chromosome and (b) the ratio of fusion events to fusion opportunities – the latter taking into account a tree structure which relative to the current investigation is a random variable. For mice, the dependent variables were (a) the number of combinations that a chromosome is involved in and (b) the number of populations in which a chromosome is in a fused state. The explanatory variables in both species were chromosome size, GC content, gene density and, additionally for the mouse, HKP gene density. Fusion of high-HKP chromosomes has been hypothesized to be selected against as it may influence the regulation of these genes (Ruiz-Herrera et al., Reference Ruiz-Herrera, Farré, Ponsà and Robinson2010). GC content and gene density are included since they may mediate chromosome positioning (GC content suggesting itself as a plausible part of a mechanism by which gene density-based positioning could be effected). Chromosome size is included for its observed relationship with Rb fusion frequency (Gazave et al., Reference Gazave, Catalan, da Graça Ramalhinho, da Luz Mathias, Nunes, Dumas, Britton-Davidian and Affray2003) and chromosome positioning. We tested (using available data on chromosomes 1, 2, 9, 11 and 14) whether the mouse chromosome positioning in any of four cell type/phase combinations (lymphocyte S phase, fibroblast S phase, embryonic stem cell S phase and macrophage G0 phase) or the average position across cell types monitored in S-phase or across all cell types, can predict Rb fusion frequency.
(v) Partitioning autosomes into groups by size
In the case of bovids, the autosomes were divided into three groups by size based on natural size gaps between chromosomes 11 and 12 and then chromosomes 17 and 18, with chromosome 20 assigned to the medium group due to its relatively large size (Table 2).
Table 2. Summary data on bovid chromosome groupings used

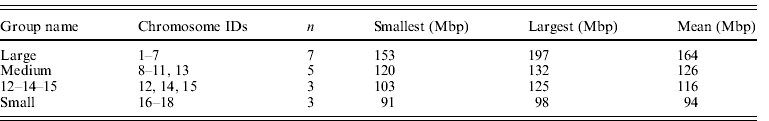
The size groupings of mouse autosomes follow Parada et al. (Reference Parada, McQueen, Munson and Misteli2002), Gazave et al. (Reference Gazave, Catalan, da Graça Ramalhinho, da Luz Mathias, Nunes, Dumas, Britton-Davidian and Affray2003) and Ruiz-Herrera et al. (Reference Ruiz-Herrera, Farré, Ponsà and Robinson2010) . Chromosome 19 was omitted as a known extreme value (it is the smallest chromosome by a considerable margin) although it is acknowledged that it fits general trends (Gazave et al., Reference Gazave, Catalan, da Graça Ramalhinho, da Luz Mathias, Nunes, Dumas, Britton-Davidian and Affray2003). Chromosomes 12, 14 and 15 were assigned to their own group (group 3) as they have been suggested to occupy a territory by themselves (Parada et al., Reference Parada, McQueen, Munson and Misteli2002). This left the large, medium and small groups shown in Table 3. Alternative groupings ((1–7, 8–15, 16–19) and (1–7, 8–12, 13–18)) did not differ from the one favoured here.
Table 3. Summary data on mouse chromosome groupings used
(vi) Correction for chromosomal combinatorics
To obtain comparable values for visualizing the data, it was necessary to take into account that different size groups have different number of members, e.g., there were 11 chromosomes in the bovid ‘large chromosome’ group, and seven in the bovid ‘medium-sized chromosome’ group. Homotypic fusions (where both participating chromosomes are from the same size-group) have a probability space of ni (ni −1)/2, where ni is the number of chromosomes in the group, whereas heterotypic fusions have ni nj , with ni and nj being the potentially different numbers of chromosomes in the two groups. We therefore divided occurrence frequencies by the size of the probability space for each combination of groups. We referred to the resulting value as ‘double-averaged’; in the case of the number of observed fusion partners in the mouse, the value reflects the observed fraction of possible partnering for each size-group.
(vii) Spindle-induced bias in the house mouse
We re-analysed Pardo-Manuel de Villena & Sapienza's (Reference llena and Sapienza2001) data in combination with those collated by Piálek et al. (Reference Piálek, Hauffe and Searle2005) to examine the suggestion that spindle polarity (the spindle reversal hypothesis) could explain Rb fusion frequencies. Analysis was by GLM, nonparametric tests, and via groups. Groups had to be defined differently here (large (L): chromosomes 1–7, medium (M): chromosomes 8–15, small (S): chromosomes 16–19) due to less data being available. We have not provided a corresponding analysis for bovids since data on spindle preferences for, or against, metacentric chromosomes are unavailable.
3. Results
(i) Chromosome size predicts Rb fusion participation in bovids
The ratio of participations over opportunities (hereafter ‘ratio’) is more highly correlated with chromosome length (Pearson's r=0·709) than participations alone (Pearson's r=0·670). A GLM fitting chromosome length as an explanatory variable is also more significant when the ratio is used (P=1·66×10−5 versus P=7·08×10−5; Fig. 1 a and b, respectively, with length consistently having P<0·0001); however, when opportunities are added as an additional variable explaining participations, opportunities drop out of the analysis (P=0·23). GC content is also marginally nonsignificant when fitted as an additional variable (P=0·0578, adjusted R 2=0·484, up from 0·428 when not fitting GC) for participations alone, but seemed to explain additional variation in the ratio (P=0·0318, adjusted R 2=0·553, up from 0·484 when not fitting GC).

Fig. 1. (a) Incidence of Rb fusions divided by fusion opportunities plotted against chromosome sizes (Pearson's c.c.=0·6915, R 2=0·4588, P=3·27×10−5). (b) Incidence of Rb fusions within bovids plotted against known chromosome sizes in B. taurus (Pearson's c.c.=0·6697, adjusted R 2=0·4281, P=7·08×10−5). (c) Mouse chromosome data for comparison, showing size versus observed number of different fusion partners. Large mouse acrocentric chromosomes fall outside the range covered by bovid acrocentric chromosomes, and fuse less frequently (Gazave et al., Reference Gazave, Catalan, da Graça Ramalhinho, da Luz Mathias, Nunes, Dumas, Britton-Davidian and Affray2003).
However, GC content taken on its own was an unsatisfactory explanatory variable (P=0·28 and 0·19 when describing participations and ratio, respectively). This suggests (a) that size, not GC content or one of its correlates (gene density, HKP genes and SINEs), is the stronger determinant of Rb fusion and chromosomal positioning, and (b) that size influences Rb fusion frequency through at least one mechanism not related to the well-understood relationship between size and GC content. We confirmed the negative correlation between chromosome size and GC content reported by previous studies (Fig. 2). The most complex model of bovid data had 25 remaining degrees of freedom; GLM analyses of bovid data are summarized in Supplementary Table S2.

Fig. 2. Genomic GC of B. taurus chromosome content plotted against chromosome size expressed in Mbp. Pearson's c.c.=−0·6249124, adjusted R 2=0·3679, P=0·00029.
(ii) Role of GC and gene density in explaining Rb fusions in mice
Given the relationship of fusion frequency with mouse chromosome size (Gazave et al. (Reference Gazave, Catalan, da Graça Ramalhinho, da Luz Mathias, Nunes, Dumas, Britton-Davidian and Affray2003), we considered GC as an explanatory variable in isolation, and with chromosome size in a GLM. In addition to testing for a linear relationship, testing a quadratic function was motivated by Kuraku et al.'s (Reference Kuraku, Ishijima, Nishida-Umehara, Agata, Kuratani and Matsuda2006) observation of a unimodal relationship between chromosome size and GC content in the mouse. After excluding two outliers from the analysis (chromosomes 11 and 19), the square of GC content was a significant predictor of the number of observed fusion pairs of a chromosome, both on its own (P=0·041 for the squared term), and with the square of chromosome size included in the model (all P values between 0·006 and 0·007). Outlier exclusion meant that the most complex model (chromosome length, GC content, gene density and HKP gene density, all as squared factors) had eight remaining degrees of freedom. The order of model preference from most to least preferred was: (1) model with both squared terms (GC content and size, adjusted R 2=0·492), (2) model with GC squared as sole predictor (adjusted R 2=0·163), followed by (3) model with chromosome size squared as sole predictor (adjusted R 2=0·148). Results of analyses aimed at predicting the number of observed chromosomal combinations are summarized in Supplementary Table S3.
When gene density was added to the most preferred model as a quadratic term, both GC content and gene density had a significant effect (P=0·031 and 0·036, respectively, for the squared term) and improved the model (adjusted R 2=0·667); gene density was also highly significant on its own (P=0·0038). However, the number of HKP genes was not a useful addition to any of the consecutively improving models, and is only marginally significant after Bonferroni correction if outliers were included (uncorrected P=0·011, adjusted R 2=0·283, otherwise P=0·48), and not in combination with other predictors. (Bonferroni correction for testing four explanatory variables – chromosome size, GC content, gene density, HKP gene density – entailed reducing the 0·05 significance thresholds to 0·0125).
As an explanation of the number of populations carrying a chromosome involved in a fusion product (i.e. as one-half of a metacentric), GC content squared was only a marginally significant predictor on its own, and was nonsignificant after Bonferroni correction (P=0·0408, correction as above). In contrast, size remained significant with P=0·00196. Including the previously suggested outliers did not improve the role of GC content in the model – in fact, including them made chromosome size more significant, possibly reflecting violations of normality. The inclusion of gene density similarly failed to improve the model. HKP gene density had a nearly significant negative effect when fitted as a linear predictor. This is consistent with an earlier finding by Ruiz-Herrera et al. (Reference Ruiz-Herrera, Farré, Ponsà and Robinson2010), and was true even though chromosome 19 (which their study was based on) was excluded as an outlier.
A possible explanation is that occurrences of fused states in different populations are not entirely independent events, but are, in at least some instances, synapomorphic. Results of analyses of the number of populations carrying given chromosomes in a metacentric state are summarized in Supplementary Table S4. Based on adjusted R 2, the preferred model has chromosome length as a quadratic term as its only predictor.
(iii) Effect of chromosome positioning on Rb fusions
Analysing a subset of chromosomes dictated by data provided by Mayer et al. (Reference Mayer, Brero, von Hase, Schroeder, Cremer and Dietzel2005) resulted in a close (nonsignificant) correspondence between chromosome positioning and (a) the number of other chromosomes seen to fuse with a particular chromosome and to a lesser extent (b) the number of populations in which such a fused state was observed (Supplementary Table S5). This weaker fit for population occurrence is consistent with shared fused chromosomes that are likely to have resulted, in at least some cases, from gene flow (i.e. synapomorphy). More importantly, estimated coefficients were consistently negative, meaning that in this subset chromosomes with more peripheral positions had lower Rb fusion participation.
The nearly significant fit between lymphocyte S-phase position of chromosomes and their participation in Rb fusions (for a number of combinations observed, P=0·108) should be treated with caution given the small sample size (n=5) and non-orthogonality of the design. Although multiple testing should enter into interpretation, it is worth noting that several tests had similarly close-to-significant results, with chromosome placement correlated between different cell types and phases (all r between 0·50 and 0·74).
(iv) Fusion preferences by size classes
We found that chromosome size classes behaved similarly with respect to preferred partners. In the case of the bovids all size classes mostly tended to partner with large chromosomes, and tended least to partner with small chromosomes (Fig. 3). This manifests as a significant linear correlation of corrected frequency (taking into account size group membership and resulting combinatorics) against the size of resulting metacentric chromosomes (P=0·0062, R 2=0·87).

Fig. 3. Double-averaged fusion frequencies for different combinations of bovid chromosomes grouped by size. There is no evidence of chromosomal territoriality in bovids based on 195 Rb fusions (see text) – small chromosomes are the least popular partner for all chromosomes, with large chromosomes being correspondingly more preferred. ‘Double averaging’ corrects for unequal group size and different combinatorics of homotypic versus heterotypic groups.
Mouse chromosomes, in contrast, behaved differently in terms of chromosome partnering (Fig. 4). The same chromosomes previously noted as sharing a single territory (specifically numbers 12, 14 and 15 as observed in splenocytes; Parada et al., Reference Parada, McQueen, Munson and Misteli2002), were prolific in obtaining fusion partners, particularly larger than themselves. Small and large chromosomes did not fuse as often – a finding consistent with the idea that CT positioning according to chromosomal sizes (e.g. Bolzer et al., Reference Bolzer, Kreth, Solovei, Koehler, Saracoglu, Fauth, Müller, Eils, Cremer, Speicher and Cremer2005) affects fusions. It is interesting that fusions of chromosomes 12–14–15 with another member from this group (i.e. Rb12·14, Rb12·15 and Rb14·15) were extremely rare – only one of the 479 occurrences of fusions in the 97 documented populations was of this kind; this is in stark contrast to fusions of the same autosomes with intermediate-sized partners being the most frequent of all combinations. This is remarkable as these two groups are closest to each other in chromosome size, suggesting it is not physical proximity per se that is preventing Rb fusions in this case. Rather, it is an additional mechanism of suppression or natural selection that may be related to territory sharing, and potentially to suppression of recombination between regions of sequence homology on different chromosomes that entitles membership of a particular, shared territory.

Fig. 4. House mouse fusion frequencies averaged for number of chromosomes in either group. (a) Fusions found in the wild. (b) Number of populations with those fusions. Based on 97 populations polymorphic for Rb fusions for a total of 479 occurrences of fusions, and 99 different fused pairs. Data from Piálek et al. (Reference Piálek, Hauffe and Searle2005) .
(v) Relationship of opportunities and participations in bovids
The available phylogeny permits an estimation of the duration that a chromosome spent fused or unfused in the history of extant bovids. We considered that if a chromosome was present in an unfused state, while other chromosomes were undergoing fusions, it had missed ‘opportunities’ for fusion. If a chromosome has allowed a significantly large number of such opportunities for fusion to lapse, this suggests either that the chromosome is less prone to undergoing fusions, or, less parsimoniously, that its early fusions did not persist (less parsimonious because fissions are thought to be relatively rare, and are almost entirely absent from the bovid dataset; Robinson & Ropiquet, Reference Robinson and Ropiquet2011). On this basis, we could consider some chromosomes to be fusion prone, while others are fusion averse.
A high number of opportunities can be taken as an indication of a chromosome's reluctance to engage in fusions, or, conversely, an indication of how ancient a particular fusion is (since having few missed opportunities requires early establishment). In other words, having fewer missed opportunities tends to indicate a higher propensity for a given chromosome to fuse. Our data show the relationship between opportunities and fusions to be significantly negative in the bovid data (P=0·0025; Fig. 5). This confirms that fusion-prone chromosomes fused sooner than fusion-averse ones, and shows that our data avoided the potentially confounding effect that there are more late branches available than early ones. Outliers from this negative relationship generally support the observed linear relationship between size and likelihood to fuse–chromosome 1 fused more often than predicted given its opportunities, whereas chromosome 22 is an outlier in the opposite direction (Fig. 5).

Fig. 5. Participations plotted against opportunities for the bovid data; Pearson's c.c.=−0·5391226, adjusted R 2=0·2644, P=0·002546.
(vi) Spindle polarity-induced bias in the house mouse
Re-analysis of Pardo-Manuel de Villena & Sapienza's (Reference llena and Sapienza2001) house mouse data of non-random segregation in combination with Piálek et al.'s (Reference Piálek, Hauffe and Searle2005) information on fusion occurrence showed an inclination for some chromosomes to segregate differently from others. The inference is made cautiously as chromosome 14 was not represented in the data, and the design (insofar as a design is inherent in a meta-study) is far from orthogonal, resulting in too few degrees of freedom to analyse all chromosomes simultaneously. Accordingly, results presented are a consensus of repeated runs of the analyses with different orders of factors. To mitigate this, we individually confirmed the suggested coefficients from the table. However, this does not altogether resolve the problems of collinearity discussed above.
Focusing only on the highly significant results in the mouse, Rb fusions involving chromosomes 4 and 6 were disfavoured, with effects of −0·21 and −0·25, respectively, on Rb transmission probability (with P-values of 0·0050 and 0·0060, respectively). Chromosome 12 had a positive effect of 0·26 at a P-value of 0·0040. Additionally, chromosomes 2 and 3 had weaker negative effects (approx. −0·1) of borderline significance. These results were obtained at the exclusion of the male sex where meiosis is known (with few exceptions) not to significantly bias. Note that in the Pardo-Manuel de Villena & Sapienza's (Reference llena and Sapienza2001) study, overall female meiotic bias was against Rb fusion products. We found the GC content of Rb fusion products correlates positively with success in segregation (P-value of 0·037), but not after Bonferroni correction (having tested for an effect of chromosome size previously, which had no significant effect).
Having established that there is some significant variation among chromosomes for their meiotic bias, we next tested whether female meiotic biases could explain departures in Rb fusion frequency of individual mouse chromosomes. Chromosomes 4 and 6 having a negative effect, and chromosome 12 a positive one, would be consistent with the observation of intermediately sized chromosomes being most frequently engaged in fusions, suggesting they might be preserved through favourable bias in female meioses. Indeed, group-based analysis showed medium-sized chromosomes (M) to be the most favoured of possible partners within each group (M with highest probability of meiotic success among pairings with S, M or L; Supplementary Figure S1 available online at http://journals.cambridge.org/grh).
We were restricted to using nonparametric tests (due to non-normality of the occurrence data) when comparing metacentric chromosome success in mouse meiosis versus population occurrence of that chromosome. Spearman's rank correlation coefficient was small and nonsignificant (r=0·26, P=0·21). Note that two metacentric variants (Rb6·18 and Rb9·19) included in Pardo-Manuel de Villena & Sapienza's (Reference llena and Sapienza2001) data had not been reported as occurring naturally by Piálek and co-workers. Exclusion of these data points did not qualitatively affect the result.
4. Discussion
Rb chromosomal polymorphisms in current-day populations of the house mouse may merely represent transient short-term evolutionary episodes (Gazave et al., Reference Gazave, Catalan, da Graça Ramalhinho, da Luz Mathias, Nunes, Dumas, Britton-Davidian and Affray2003; Piálek et al., Reference Piálek, Hauffe and Searle2005; transience in the house mouse is likely caused by extinction of metacentric populations rather than fission, Britton-Davidian et al., Reference Britton-Davidian, Catalan, da Graça Ramalhinho, Auffray, Nunes, Gazave, Searle and da Luz Mathias2005). Our study, however, offers an additional perspective on Rb fusions that have persisted for millions of years, and is therefore directly relevant to evolution in the long term, with a potential to contribute to reproductive isolation. We report that in both bovids and the house mouse, Rb fusion occurrence depends on chromosome size, GC content, and, to a lesser extent, density of genes and HKP genes. All of these are previously acknowledged correlates of the positioning of chromosomes in interphase and prometaphase nuclei, suggesting that chromosome positioning may cause the fusion patterns we see.
(i) Size and position influence fusion participation
Although no complete datasets of chromosome positioning are available for our taxa of interest, Koehler et al. (Reference Koehler, Zakhartchenko, Froenicke, Stone, Stanyon, Wolf, Cremer and Brero2009) have determined positions of chromosomes 19 and 20 in bovine embryos and Mayer et al. (Reference Mayer, Brero, von Hase, Schroeder, Cremer and Dietzel2005) of mouse autosomes 1, 2, 9, 11 and 14 in a variety of cell types. Comparison of autosomal positioning with fusion frequency led to results consistent with centrally positioned chromosomes having higher fusion frequency. The available autosomes were of a variety of GC contents, gene densities and sizes, but none smaller than chromosome 14. Hence, the analysis could not have detected the unimodal pattern previously reported for size by Gazave et al. (Reference Gazave, Catalan, da Graça Ramalhinho, da Luz Mathias, Nunes, Dumas, Britton-Davidian and Affray2003) . In contrast, we report a linear relationship for bovids, with likelihood of participation increasing with chromosome size.
Dividing chromosomes into size classes to establish whether any structure might arise from the spatial arrangement of chromosomes (where according to our physical proximity hypothesis, large chromosomes should be neighbours to medium-sized ones and medium-sized chromosomes neighbour small ones) showed such an effect in bovids where likelihood of a fusion was fairly well predicted by the length of the fused metacentric. Importantly, our partial re-analysis of Mayer et al.'s data suggested that S-phase nuclei may have similar chromosome positioning to that which is relevant when Rb fusions are formed in the germ line. Performing the size group analysis on an updated version of the mouse dataset (Piálek et al., Reference Piálek, Hauffe and Searle2005), we show that the likelihood of interaction does depend on proximity in size, likely a reflection of proximity within the nucleus (Bolzer et al., Reference Bolzer, Kreth, Solovei, Koehler, Saracoglu, Fauth, Müller, Eils, Cremer, Speicher and Cremer2005) and thus, to our knowledge, the first evidence that chromosomal location may affect the selection of chromosomes for Rb fusion in this species.
(ii) Other genomic correlates
Having assessed the effects of several genomic variables on Rb frequency, it is worth considering whether these are truly orthogonal, and how this may affect the differences observed between mouse and bovids. The published literature makes it clear that these variables are related by a number of mechanisms (Supplementary Fig. S2). Parts of this causal network are complex, i.e. the feedback relationship between GC content and rate of recombination. In other cases, the predicted trend is unclear, as in the relationship of chromosome size and GC content which is inversely correlated in humans and cattle (Fig. 2), but not the mouse (Kuraku et al., Reference Kuraku, Ishijima, Nishida-Umehara, Agata, Kuratani and Matsuda2006). The mouse's unusual pattern of GC content with chromosome size may be due to its shorter generation times. This may have allowed molecular evolution to progress further in the mouse than many other mammals (Romiguier et al., Reference Romiguier, Ranwez, Douzery and Galtier2010) such that the opposing forces of GC accumulation by gene conversion and simultaneous GC erosion in the GC-richest isochores (Duret et al., Reference Duret, Semon, Piganeau, Mouchiroud and Galtier2002; Arndt et al., Reference Arndt, Petrov and Hwa2003; Belle et al., Reference Belle, Duret, Galtier and Eyre-Walker2004) have progressed to a stage beyond that evidenced by bovids. Therefore, the absence of a strong GC content effect in determining population persistence of Rb fusions in the mouse should not lead to conclusions that this is likely true for other mammals.
(iii) Large chromosomes fuse most frequently in bovids
We found that bovid chromosomes significantly preferred to partner with large chromosomes and avoided partnering with small chromosomes (Fig. 3). Under prevailing CT theory, large chromosomes should mostly be found on the periphery of the nucleus and small chromosomes at the centre (Bolzer et al., Reference Bolzer, Kreth, Solovei, Koehler, Saracoglu, Fauth, Müller, Eils, Cremer, Speicher and Cremer2005). This would confer low connectivity on large chromosomes, making them fuse less often. The alternate model of freely intermingling chromosomes, devoid of territorial associations, suggests no such disadvantage. In fact, large chromosomes would have a larger ‘catchment area’ available to initiate fusions.
The first step in generating Rb fusions is currently believed to be the creation of a double-stranded break (DSB; e.g. Dumitrache et al., Reference Dumitrache, Hu and Hasty2009). For what are called non-recurrent Rb fusions, these DSBs would have to occur on the small p-arms of the bovid acrocentrics in order to facilitate Rb fusion (e.g. Dumitrache et al., Reference Dumitrache, Hu and Hasty2009). The five acrocentric human chromosomes 13, 14, 15, 21 and 22 show a trend for smaller acrocentric chromosomes to have absolutely shorter p-arms (data in Wise et al., Reference Wise, Crout, McNeil, Weyant, Marazita and Wenger2009; correlation between length of p-arm and q-arm: P=0·023, R 2=0·86), suggesting that larger chromosomes would indeed make a larger target for initiation of Rb fusions; the size of the telomere similarly scales with arm length (Wise et al., Reference Wise, Crout, McNeil, Weyant, Marazita and Wenger2009). Although telomeres typically vary by ~15 kbp between the long and short arm (not enough to explain the up to 1 Mbp difference in short arm length among acrocentrics), longer telomeres are also associated with greater resistance to fusion (Slijepcevic et al., Reference Slijepcevic, Hande, Bouffler, Lansdorp and Bryant1997 and references therein). Therefore, while the increased length of the short arm suggests a mechanism by which an increased incidence of non-recurrent fusion, mediated by longer short arm targets for DSBs, might explain the bovid data, the actual outcome would depend on the strength of the suppression effect created by the relatively larger telomeres. The appeal of such an explanation lies in it nullifying the need for direct natural selection and/or CTs. The fact that there is individual variation in telomere length (e.g. Treff et al., Reference Treff, Su, Taylor and Scott2011) suggests that telomere length may be positively selected for in chromosomes whose participation in fusion is selected against, with weaker selection and loss of telomere units affecting chromosomes whose fusion is less unfavourable.
It is also interesting that the stronger resistance to fusion caused by the large telomeres typical of large chromosomes would counteract the observed trend, in that if telomeres were the same length for all chromosomes, the difference between large and small in fusion frequency would be even greater in bovids. Consequently, while we have good evidence on the variation in length of telomeric TTAGGC repeats, identification of the identity of recurrent Rb-anchoring sequences has yet to be followed by detailed studies of their size variation.
Prophase of meiosis I is believed to be the main source of DSBs (Scherthan et al., Reference Scherthan, Weich, Schwegler, Heyting, Härle and Cremer1996; Ashley et al., Reference Ashley, Gaeth, Inagaki, Seftel, Cohen, Anderson, Kurahashi and Emanuel2006), and a checkpoint exists in metaphase I that biases against imbalanced meioses, and possibly against Rb fusions (Eaker et al., Reference Eaker, Pyle, Cobb and Handel2001; Manterola et al., Reference Manterola, Page, Vasco, Berríos, Parra, Viera, Rufas, Zuccotti, Garagna and Fernández-Donoso2009). It is thought that prophase I mostly produces ‘programmed’ DSBs (Ashley et al., Reference Ashley, Gaeth, Inagaki, Seftel, Cohen, Anderson, Kurahashi and Emanuel2006). Programmed DSBs are believed to improve the accuracy of homologous chromosome segregation, and are the main or exclusive source of ‘recurrent’ chromosomal translocations (Ashley et al., Reference Ashley, Gaeth, Inagaki, Seftel, Cohen, Anderson, Kurahashi and Emanuel2006). Recurrent translocations are those that have been found to independently result from the same breakpoints multiple times. These breaks occur in repetitive DNA on different chromosomes and can produce Rb fusions (Rowley & Pergament, Reference Rowley and Pergament1969; Hecht & Kimberling, Reference Hecht and Kimberling1971; Choo et al., Reference Choo, Vissel, Brown, Filby and Earle1988; Nanda et al., Reference Nanda, Schneider-Rasp, Winking and Schmid1995; Page et al., Reference Page, Shin, Han, Choo and Shaffer1996; Bandyopadhyay et al., Reference Bandyopadhyay, Berend, Page, Choo and Shaffer2001). A typical breakpoint location is within pericentromeric satellite sequence (Nanda et al., Reference Nanda, Schneider-Rasp, Winking and Schmid1995; Page et al., Reference Page, Shin, Han, Choo and Shaffer1996; Bandyopadhyay et al., Reference Bandyopadhyay, Berend, Page, Choo and Shaffer2001; Adega et al., Reference Adega, Guedes-Pinto and Chaves2009; Gauthier et al., Reference Gauthier, Hima and Dobigny2010). A large proportion of mammalian Rb fusions are recurrent in this sense (Nanda et al., Reference Nanda, Schneider-Rasp, Winking and Schmid1995; Bandyopadhyay et al., Reference Bandyopadhyay, Berend, Page, Choo and Shaffer2001), and therefore highly likely to stem from meiotic prophase I, where explicit allowance is made by cellular mechanisms for the required DSBs. Considerable data now support the idea that homologous chromosomes find each other at a time in early prophase during which telomeres are being dragged along the nuclear envelope (Harper et al., Reference Harper, Golubovskaya and Cande2004). Although not essential, this motion speeds up chiasma formation and development of the synaptonemal complexes (Harper et al., Reference Harper, Golubovskaya and Cande2004). However, this same increase in kinetic energy may also increase the incidence of DSBs. This would argue for a leptotene/zygotene timing of the majority of Rb fusions. For fusion events that occur prior to metaphase checks, there is the potential that longer chromosomes will be delayed and have fewer DSBs correctly resolved, and more Rb fusions retained.
(iv) Medium-sized chromosomes fuse most frequently in the house mouse
The typical observation of nuclear architecture has been one of the radial arrangements where large chromosomes are found on the periphery of the nucleus and smaller chromosomes at the centre (Cremer et al., Reference Cremer, von Hase, Volm, Brero, Kreth, Walter, Fischer, Solovei, Cremer and Cremer2001 and references therein). This radial arrangement somewhat fits a previous observation that M. musculus chromosomes of intermediate size are most frequently found to be involved in Rb fusions (Gazave et al., Reference Gazave, Catalan, da Graça Ramalhinho, da Luz Mathias, Nunes, Dumas, Britton-Davidian and Affray2003), suggesting that chromosomes of intermediate size might have an opportunity to fuse with both peripheral as well as less peripheral chromosomes.
The particular pattern in the mouse suggests that interactions predominantly occur along radial gradients. Only a small subset of all the possible combinations of a small chromosome fused with another small chromosome, or as a combination of small chromosomes fused with either chromosome 12, 14 or 15, was observed. This might indicate that there is a barrier to interactions at the centre of the radial pattern (perhaps associated with the nucleolus). On this point at least, the bovid and mouse results are consistent with each other, although the causes may be different.
Another possibility is that chromatin organization is more constrained at the more gene dense, short chromosome-containing centre of the nucleus, thus permitting relatively fewer interchromosomal translocations than the abundance of neighbours would imply. In support of this, there would be a higher concentration of euchromatin and associated signals, promoters and enzymes (including ones required for DSB repair). The higher concentration would be expected simply as a result of having a higher concentration of binding targets at the euchromatin-rich centre of the nucleus and in this regard, differential width of the ICD may be important. Interestingly, the observed pattern of high fusion participation of medium-sized mouse chromosomes closely matches radial CT distances in flat-ellipsoid human fibroblast interphase nuclei and mitotic prometaphase rosettes (Fig. 3 in Bolzer et al., Reference Bolzer, Kreth, Solovei, Koehler, Saracoglu, Fauth, Müller, Eils, Cremer, Speicher and Cremer2005) as well as spherical human lymphoblast nuclei (Fig. 2 in Boyle et al., Reference Boyle, Gilchrist, Bridger, Mahy, Ellis and Bickmore2001), where the largest chromosomes were less peripheral in position than medium-sized ones.
(v) Possible role of spindle polarity-induced bias in the house mouse
We tested for a possible role of spindle bias in explaining our results for the house mouse as was suggested by Pardo-Manuel de Villena & Sapienza (Reference llena and Sapienza2001) . There was evidence that spindle bias may be correlated with GC content and, based on an incomplete dataset, that spindle bias seemed to promote fusion of particular chromosomes, supporting the notion of an advantage for intermediate-sized chromosomes. Rather than attempt to compute chromosome-by-chromosome bias values and compare these to the number of metacentric pairings in which a chromosome is found in the wild, we made a direct comparison of the meiotic bias observed for a particular metacentric with the abundance of that metacentric in wild populations. The corresponding Spearman's rank correlation coefficient was small and nonsignificant. However, when analysed by groups, there is a clear pattern of the dominant spindle pole (i.e. the one which, with its surrounding cytoplasm, continues on towards the egg cell fate) in meiosis I favouring medium-sized chromosomes, such that for any chromosome to succeed in segregation after fusion, partnering with a medium-sized chromosome is best. This pattern closely matches the grouped frequencies of mouse chromosome Rb fusion, suggesting that spindle biases are at least consistent with the Rb frequencies. We would therefore cautiously suggest that the abundance of Rb races in the house mouse may either not require reversals in spindle polarity, in contrast to what was previously assumed (Pardo-Manuel de Villena & Sapienza, Reference llena and Sapienza2001), or that the weak meiotic bias in favour of medium-sized chromosomes is independent of spindle polarity. This suggests that spindle reversal may be necessary to create a climate in which Rb fusions persist more frequently, but that the persistence of some Rb fusions more than others is due to a different mechanism and cause.
(vi) Concluding comments
In both bovids and the house mouse, chromosome size is the major determinant of a chromosome's likelihood to undergo Rb fusion. However, there is a remarkable difference between the two datasets in how chromosomes ‘network’. This suggests that either low-level molecular processes in the mammalian nucleus are more taxonomically diverse than previously thought, or that the observed outcome is sensitive to the timescale investigated, with the Rb fusions that differ from one species to another being qualitatively different from population-level polymorphisms that change on relatively short timescales (Britton-Davidian et al., Reference Britton-Davidian, Catalan, da Graça Ramalhinho, Ganem, Auffray, Capela, Biscoito, Searle and da Luz Mathias2000). Many of the fusions in bovids occurred on recent branches (Robinson & Ropiquet, Reference Robinson and Ropiquet2011). This gives some support to the idea that the patterns of Rb fusion differ in a substantial way between murids and bovids even when timescales involved are similar, possibly via less enforced territories during meiotic prophase I in bovids.
The mouse data support the physical proximity hypothesis, but other weak evidence points at a contribution from meiotic bias, possibly via a spindle-related mechanism. Although data on the positioning of the complete set of chromosomes in either target taxon are still missing, it is noteworthy that the same factors that have been found to affect the positioning of chromosomes also explain most of the variation in Rb fusion participation of those chromosomes. This suggests that spatial organization within the nucleus can indeed affect the evolutionary interactions of chromosomes. However, due to the complexities of interactions at that fine scale, further molecular work will be needed to understand exactly how particular Rb fusions are formed.
P. L. W. is a postdoctoral fellow in the Evolutionary Genomics Group. T. J. R.’s research was supported by the South African National Research Foundation. We thank Anne Ropiquet, Janice Britton-Davidian and two anonymous reviewers for comment. Lutz Froenicke critically read various versions of this manuscript. Alan Christoffels and Johann Rohwer assisted with acquiring sequence data.
5. Supplementary material
A supplementary figure, Figure S1, and five supplementary tables, Table S1–S5, are available online at http://journals.cambridge.org/grh.
Declaration of Interest
None.












