Highlights:
• STAT1 and NTD mutants can be stably expressed in U3A cells
• Similar effects of wild-type and mutant STAT1 on proliferation ± IFNα
• STAT1FA accentuates ɣH2AX accumulation and apoptosis upon ɣ-irradiation
Introduction
Binding of cytokines and growth hormones to cognate receptors activates the inducible transcription factors STAT1,-2,-3,-4,-5A,-5B,-6. STATs have a modular structure (NTD, coiled-coil, DBD, linker, SH2-domain, p-tyrosine site, C-terminal activation domain) (Khodarev et al., Reference Khodarev, Roizman and Weichselbaum2012; Wieczorek et al., Reference Wieczorek, Ginter, Brand, Heinzel and Krämer2012). STAT1 phosphorylation at Y701 leads to high affinity interactions with other STATs. The NTD allows cooperative STAT1-STAT1/STAT1-STAT2 DNA-binding of pre-formed dimers and enables STAT1 dephosphorylation (Mertens et al., Reference Mertens, Zhong, Krishnaraj, Zou, Chen and Darnell2006; Wieczorek et al., Reference Wieczorek, Ginter, Brand, Heinzel and Krämer2012).
IFN-/STAT1-related signatures promote resistance to chemotherapy and ɣ-irradiation in cancer cells (Ah-Koon et al., Reference Ah-Koon, Lesage, Lemadre, Souissi, Fagard, Varin-Blank, Fabre and Schischmanoff2016; Kaowinn et al., Reference Kaowinn, Jun, Kim, Shin, Hwang, Kim, Shin, Kaewpiboon, Jeong, Koh, Krämer, Johnston and Chung2017; Khodarev et al., Reference Khodarev, Roizman and Weichselbaum2012; Malilas et al., Reference Malilas, Koh, Kim, Srisuttee, Cho, Moon, Yoo, Oh, Johnston and Chung2013). Intriguingly, STAT1 can though promote sensitivity to DNA crosslinking agents (Prieto-Remon et al., Reference Prieto-Remon, Sanchez-Carrera, Lopez-Duarte, Richard and Pipaón2013) and pro-apoptotic gene expression (Khodarev et al., Reference Khodarev, Roizman and Weichselbaum2012; Wieczorek et al., Reference Wieczorek, Ginter, Brand, Heinzel and Krämer2012). Hence, modulators of STAT1 may be novel and innovative context-dependent anti-cancer drugs (Khodarev et al., Reference Khodarev, Roizman and Weichselbaum2012).
Objective
It is unclear whether the STAT1 NTD and DBD modulate cellular responses to fibrosarcoma cell growth and their responses to IFNα and ɣ-irradiation. We set out to answer these questions. We reconstituted STAT1 negative U3A fibrosarcoma cells with wild-type STAT1 and NTD/DBD mutants thereof. We investigated cellular responses and molecular parameters.
Material and Methods
Cell lines
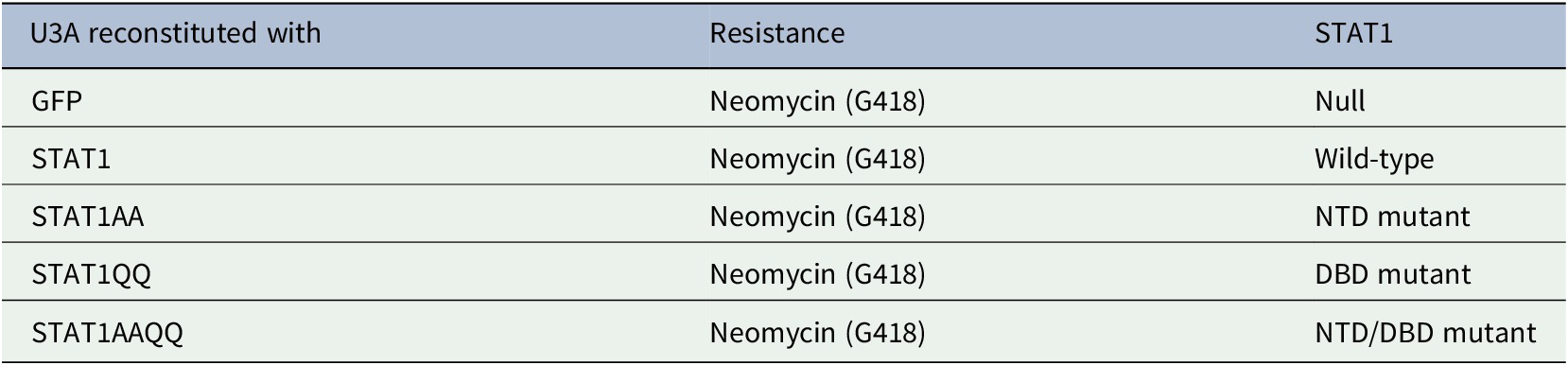
Cells were cultured as described (Ginter et al., Reference Ginter, Bier, Knauer, Sughra, Hildebrand, Münz, Liebe, Heller, Henke, Stauber, Reichardt, Schmid, Kubatzky, Heinzel and Krämer2012) and table 1:. See figure legends for ɣ-irradiation.
Plasmids
We refer to HA-tagged STAT1α as STAT1. STAT1AA has moieties phenylalanine-77 and leucine-78 replaced by alanine moieties (mutagenic primers 5-GCTTTTCTTTGGAGAATAACGCCGCGC TACAGCATAACATAAGG-3/5-CCTTATGTTATGCTG TAGCGCGGCGTTATTCTCCAAAGAAAA GCG-3). STAT1AAQQ carries these mutations and additional lysine-410 and lysine-413 exchanges to glutamine residues (Ginter et al., Reference Ginter, Bier, Knauer, Sughra, Hildebrand, Münz, Liebe, Heller, Henke, Stauber, Reichardt, Schmid, Kubatzky, Heinzel and Krämer2012).
Western blot, immunoprecipitation, antibodies, drugs, chemicals, cytokines
For Western blot technique see (Beyer et al., Reference Beyer, Kiweler, Mahboobi and Krämer2017). Antibodies: Santa-Cruz-Biotechnology (β-actin/sc-47,778;p53/sc-81,168;IFNAR/sc-845; STAT1/sc-346/sc-417; p-STAT1/sc-7,988-R); Sigma (tubulin-α/T5168); Abgent (UBCH8/AP2118b); BD-Pharmingen (cleaved PARP1/552596); Abcam (ATM/ab32420;p-S1981-ATM/ab81292); Cell-Signalling (ATR/2790;CHK1/2360;p-S317-CHK1/2344;CHK2/2662;p-T68-CHK2/2661;p-S15-p53/9284;γH2AX/9718); Millipore (53BP1/NAB3802). For immunoprecipitation, drugs, chemicals, and cytokines see (Ginter et al., Reference Ginter, Bier, Knauer, Sughra, Hildebrand, Münz, Liebe, Heller, Henke, Stauber, Reichardt, Schmid, Kubatzky, Heinzel and Krämer2012). Immunoblot data show results from at least two independent experiments.
Cell cycle and apoptotic DNA fragmentation analyses
Cells were incubated with 103 U/ml IFNα or 10 Gy using a Gammacell 2000 irradiator. Adherent and floating cells were collected. Single-cell suspensions in PBS were centrifuged (5 min, 700xg, RT). Pellets were fixed (1 ml ice cold 70% ethanol), stored at −20 °C overnight, and washed in 1 ml PBS (5 min at 700xg, RT). Per sample, 390 μl PBS, 5 μl RNase A (10 mg/ml), and 5 μl PI (2.5 mg/ml) were added (20 min, 37 °C). Samples were analyzed by flow cytometry.
Results
STAT1 and NTD/DBD mutants do not impair growth
We investigated if IFNα-activated STAT1 has anti-proliferative effects in U3A cells. STAT1 (WT) and STAT1AA (lacks NTD for dimerization and cooperative DNA binding (Mertens et al., Reference Mertens, Zhong, Krishnaraj, Zou, Chen and Darnell2006)) bound to the IFNα-receptor (IFNAR) (Fig. 1A). However, IFNα did not affect the cell cycle of U3A cells reconstituted with STAT1 or NTD (AA)/DBD (QQ) mutants (Table 1: and Fig. 1B-D).
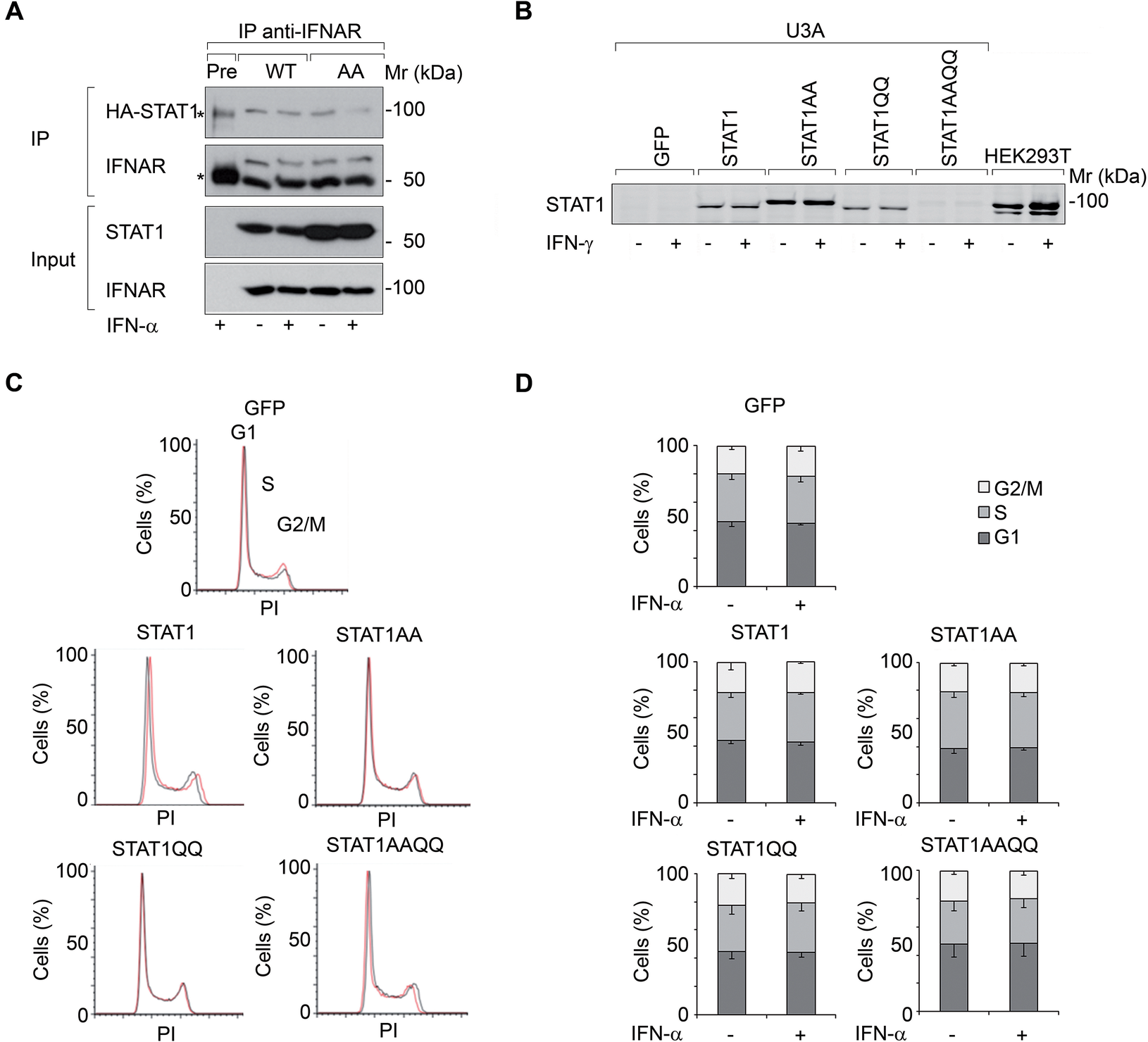
Figure 1. IFNs do not restrict the growth of fibrosarcoma cells ± STAT1.
ɣ-irradiation differentially affects U3A cells with STAT1 and its mutants
γ-irradiation evoked the accumulation of a subG1 fraction (Fig. 2A) and of cleaved PARP1 more pronouncedly in cells with STAT1AA and STAT1AAQQ (Fig. 2B). Apparently, ɣ-irradiation causes apoptosis and STAT1 accentuates this.
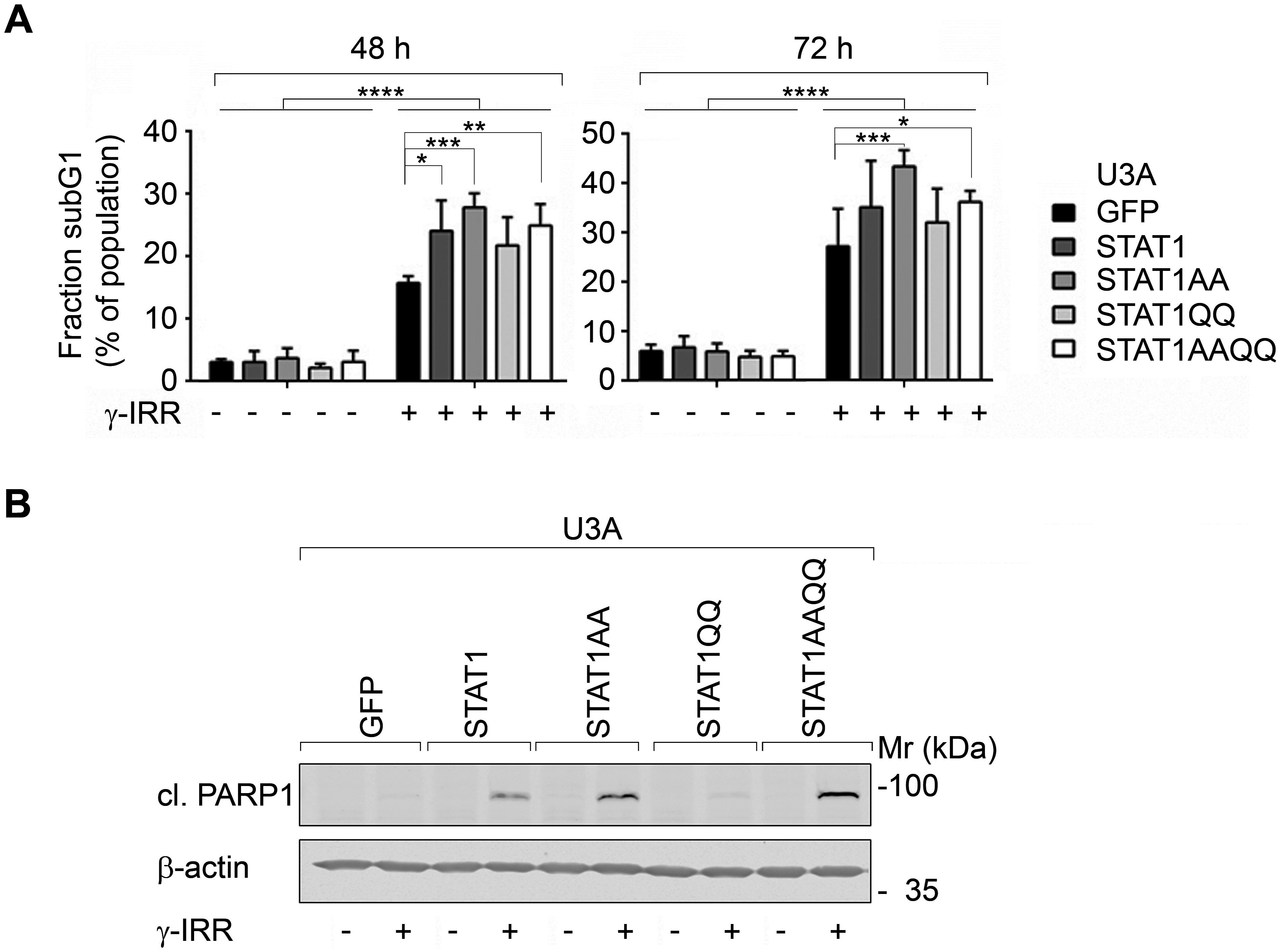
Figure 2. STAT1 sensitizes fibrosarcoma cells to cytotoxic effects of γ-irradiation.
STAT1 NTD affects ɣH2AX accumulation upon ɣ-irradiation
Next, we analyzed DNA damage sensors. 53BP1 levels were not correlated with STAT1 per se (Fig. 3A). Apical (ATM/ATR) and downstream checkpoint kinases (CHK1/CHK2) and their target p53 were phosphorylated after ɣ-irradiation irrespective of STAT1 (Fig. 3B, C). In contrast, the checkpoint kinase target ɣH2AX accumulated most pronouncedly in U3A cells with STAT1AA/STAT1AAQQ (Fig. 3C). Hence, NTD mutant STAT1 expression correlates with irradiation-induced ɣH2AX.
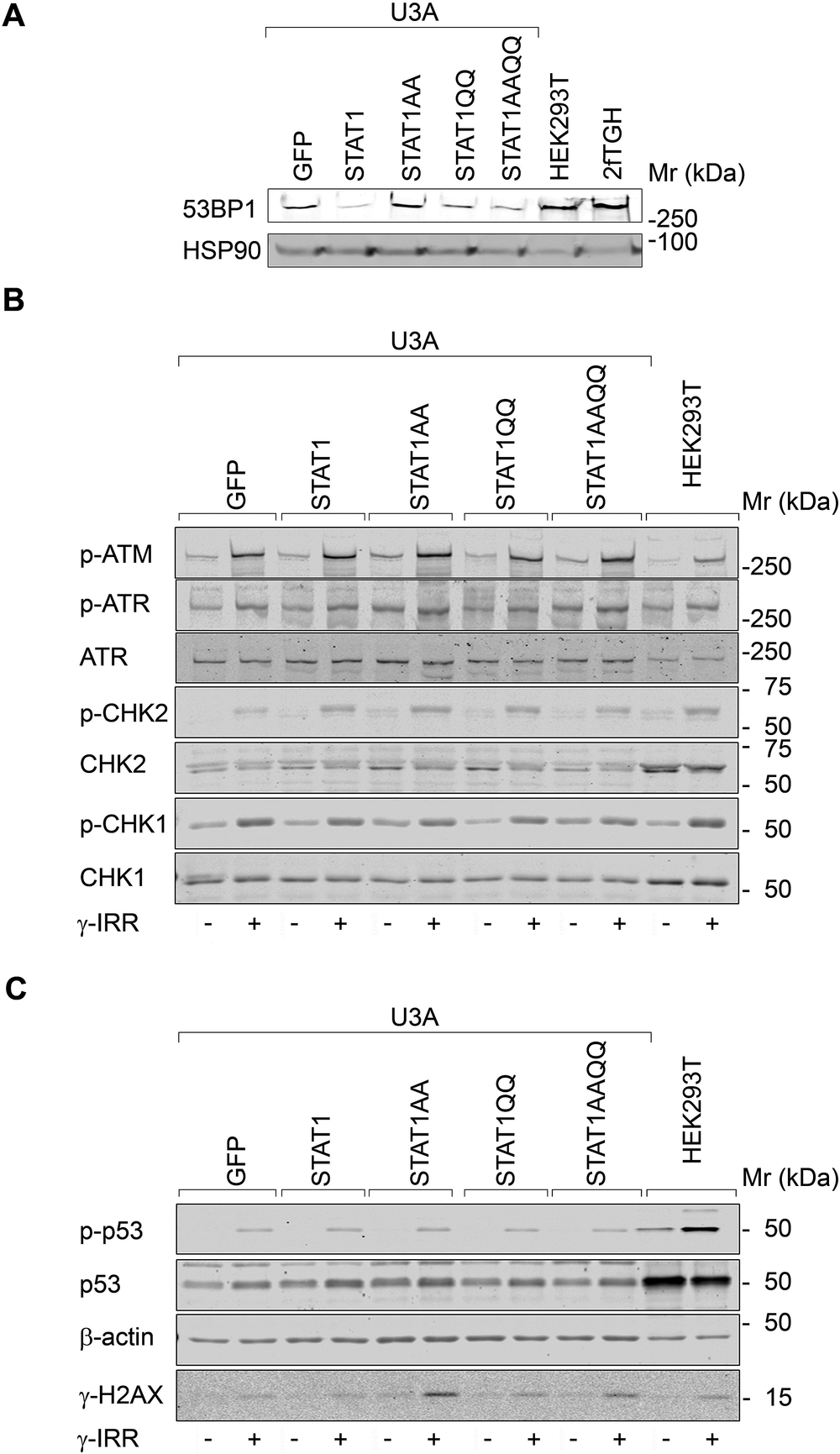
Figure 3. STAT1 does not alter checkpoint kinase signaling after ɣ-irradiation.
Discussion
STAT1 sensitizes fibrosarcoma cells to DNA damage and apoptosis upon ɣ-irradiation. Cooperativity through the STAT1 NTD seems to attenuate this. Thus, STAT1 signaling could be analyzed further as a target in fibrosarcoma and potentially other cancers. Fibrosarcoma is a clinically unmet problem with poor survival due to resistance to chemotherapy and irradiation as well as tumor recurrence after surgery. Novel treatments and the identification of mechanisms that regulate the therapeutic sensitivity of fibrosarcoma could solve the clinical problem (Augsburger et al., Reference Augsburger, Nelson, Kalinski, Udelnow, Knösel, Hofstetter, Qin, Wang, Gupta, Bonifatius, Li, Bruns and Zhao2017). Perhaps, STAT1 is both a clinically applicable marker and novel agents targeting the STAT1 NTD might give therapeutic benefits.
Conclusion
Our work provides fresh information on the relevance of STAT1 and a limited number of NTD/DBD mutants thereof for cellular responses to IFNα and ɣ-irradiation. We speculate that the higher levels of ɣH2AX in U3A cells with STAT1AA/AAQQ result from attenuated DNA repair and/or apoptosis. It is plausible that the non-cooperative STAT1 NTD mutants have binding partners that are not engaged by the dimerization-competent wild-type STAT1. Future studies are needed to reveal if such proteins might be DNA repair proteins and/or transcription factors.
Acknowledgement
We thank Sigrid Reichardt for technical assistance and Prof. Dr. T. Heinzel for helpful discussions. Moreover, we are indebted to Prof. Dr. Darnell for the HA-STAT1 plasmid.
Funding Information
Funding was from intramural funds (University Medical Center Mainz and CMB Jena).
Conflicts of interest
All authors (A.G., T.G., U.K., C.K., OH.K.) declare no conflicts of interest.
Authors contributions.
A.G., T.G., U.K. conducted data gathering.
A.G., C.K. performed statistical analyses.
A.G., O.H.K. conceived and designed the study.
OH.K. wrote the article.
Data availability statement
The data that support the findings of this study are available from the corresponding author, O.H.K., upon reasonable request.








Comments
Comments to the Author: The manuscript represents a congruent set of experimental results. There are some minor issues that should be addressed prior to acceptance:- Introduce STAT1AAQQ in the methods section- Fig. 1D. Indicate the number of biological/technical repeats and the statistical test employed- indicate the number of biological repeats performed for the WB analyses shown (e.g. “The results are representative of n biological repeats”)