Introduction
Salmonellosis is the most common bacterial cause of foodborne illness and causes approximately 1 million infections and 400 deaths annually in the USA [Reference Scallan1]. Salmonellosis is also the second most common cause of foodborne outbreaks in the USA, and the majority of these outbreaks were attributed to meat and produce [2]. However, from 1998 to 2015, there were 18 outbreaks of Salmonella infections attributed to fish, seven of which were attributed to tuna [Reference Barrett3]. The largest of these was a multistate outbreak in 2012 linked to tuna imported from India, which was served raw in sushi and resulted in 425 illnesses [4, Reference Hoffmann5].
The USA is the world's second largest seafood importer and imports an estimated 90% of the seafood eaten in the USA, most of which is fresh or frozen. Most of this seafood is imported from China, Canada and Indonesia [Reference Van Voorhees, Alan and Liddel6]. The quantity of edible seafood imported into the USA has been increasing annually, from about 2.4 million metric tons in 2011 to 2.6 million metric tons in 2015 [Reference Van Voorhees, Alan and Liddel6]. This increase is likely due to increased demand, as the estimated per capita consumption of fresh or frozen seafood in the USA has increased from 10.9 to 11.5 pounds in the same time period [Reference Van Voorhees, Alan and Liddel6].
The first known sushi restaurant opened in the USA in the 1960s, but it was not until the 1990s that sushi became a widely consumed cuisine [Reference Miyake7, Reference Avery8]. Between 1988 and 1998, the number of sushi restaurants in the USA quintupled [Reference Wine9], and by 2010 an estimated 20 000 tons of tuna were consumed annually as sushi or sashimi in the USA [Reference Miyake7]. Yellowfin tuna imports to the USA have tripled since 1989, 50–60% of which is designated for the raw tuna market [Reference Miyake7]. As sushi consumption has become increasingly popular, the USA has emerged as the second largest market for raw tuna imports behind Japan [Reference Miyake7, 10]. Combined with a decrease in the domestic supply of tuna, imports of raw tuna exceeded domestic production in the USA for the first time in 2006 [Reference Miyake7, 10].
This paper reports the results of a collaborative multistate outbreak investigation of salmonellosis involving local and state public health and regulatory agencies, the US Food and Drug Administration (FDA), and the US Centers for Disease Control and Prevention (CDC), which implicated imported frozen raw tuna served in sushi as the source of a large outbreak of Salmonella Paratyphi B variant L(+) tartrate(+) and Salmonella Weltevreden infections.
Methods
Outbreak detection
As part of routine public health activities, public health laboratories throughout the USA receive Salmonella isolates from clinical laboratories for serotyping and subtyping by pulsed-field gel electrophoresis (PFGE) [11]. These PFGE patterns are uploaded to PulseNet, the national molecular subtyping network for foodborne disease surveillance and monitored to detect illness clusters.
In April 2015, PulseNet identified a cluster of Salmonella Paratyphi B variant L(+) tartrate(+) (Salmonella Paratyphi B (dT+)) infections with an indistinguishable PFGE XbaI restriction enzyme pattern (JKXX01.1495), which had never been seen before in the PulseNet national database. State and local health departments and CDC initiated a multistate investigation to determine the source of the outbreak and prevent additional illnesses.
Case definition and case finding
We defined an outbreak-associated case as an infection with Salmonella Paratyphi B (dT+) PFGE XbaI pattern JKXX01.1495 or JKXX01.1516 in a person with illness onset between 1 March 2015 and 31 July 2015. During the investigation, two isolates with a second, similar Salmonella Paratyphi B (dT+) PFGE pattern (JKXX01.1516), which had one additional band at 30 kb compared with PFGE pattern JKXX01.1495 and was also novel to the PulseNet database, were linked to the outbreak through additional microbiologic testing and subsequently added to the initial case definition. New cases were identified through the review of clinical isolates uploaded to the national PulseNet database.
Epidemiological investigation
Local and state health departments interviewed case-patients using state-specific salmonellosis questionnaires. Based on exposure information reported from initial interviews, investigators at the California Department of Public Health developed a more detailed hypothesis-generating questionnaire and a subsequent focused questionnaire, which was used in the multistate investigation. We compared food exposure frequencies to frequencies reported in the 2006–2007 FoodNet Population Survey [12], which summarises a variety of food exposures reported by healthy people in the week before a telephone interview. To help identify exposures of interest, we compared food histories using a binomial probability distribution to determine which food exposures reported by case-patients were significantly higher than those reported by healthy people.
Traceback investigation and product testing
Local and state investigators and regulatory agencies worked with the FDA to collect invoices and conduct traceback investigations for suspected food items. Traceback is the process of tracing a food from a point of service, such as a restaurant or grocery store, to its origin or manufacturer to identify whether there is a common source for food eaten by ill people in an outbreak. Investigators used the following criteria to identify candidates for traceback: points of service associated with an illness sub-cluster (i.e. two or more case-patients with exposure to the same point of service), case-patients with a single or limited exposure to the suspected food vehicle, or case-patients with good recall of the foods consumed before illness and/or receipts documenting their food purchases.
Local and state health and regulatory agencies conducted investigations at points of service where case-patients reported eating foods of interest identified during hypothesis generation. Investigators collected restaurant menus, credit card receipts and recipes or ingredient lists for reported foods. We reviewed menu items and ingredient lists for common ingredients. Local and state investigators collected samples of suspected food products from points of service and distribution centres for Salmonella testing.
Laboratory investigations
State public health laboratories serotyped and subtyped clinical Salmonella isolates by PFGE using standard methodology [11]. State public health laboratories and CDC's enteric disease laboratory further characterised a subset of clinical isolates by whole-genome sequencing (WGS). The sequence data were generated with Illumina sequencing platforms (Table S1) and all genomes passed the basic quality metrics for PulseNet Salmonella WGS of raw sequence data of average Q-score >30 in both reads and at least 30× average coverage [Reference Katz13]. Genomes were then trimmed using CG-pipeline and analysed using Lyve-SET version 1.1.4f with phages masked, 20× coverage, 95% read support and positions no less than 5 bp apart [Reference Katz13]. For Salmonella Paratyphi B variant L(+) tartrate(+), hqSNP analysis was performed with 2015K-0310 assembled and used as internal reference; 2015AM-0987 was used as an outgroup and was not considered part of the outbreak. For Salmonella Weltevreden, hqSNP analysis was performed with 2014K-0912 closed reference genome. State public health and regulatory laboratories and FDA laboratories tested food products for Salmonella and further characterised any Salmonella isolates identified by serotype, PFGE and WGS. The tuna isolates sequenced was deposited in the GenomeTrakr database, which is part of the NCBI Pathogen detection website where both CDC, FDA and USDA FSIS draft genomes are all available for comparison [Reference Allard14].
Results
Salmonella paratyphi B (dT+) outbreak description
We identified a total of 65 cases of Salmonella Paratyphi B (dT+) (63 cases with PFGE JKXX01.1495 and two cases with PFGE JKXX01.1516) from 11 states: Arizona (11), California (36), Illinois (one), Michigan (two), Minnesota (four), Mississippi (one), New Mexico (six), South Dakota (one), Virginia (one), Washington (one) and Wisconsin (one) (Fig. 1). Illness onset dates ranged from 1 March 2015 to 20 July 2015 (Fig. 2). Case-patients ranged in age from <1 to 83 years (median 32 years), and 54% (35/65) were male. Eighteen per cent (11/62) of case-patients were hospitalised, and no deaths were reported.
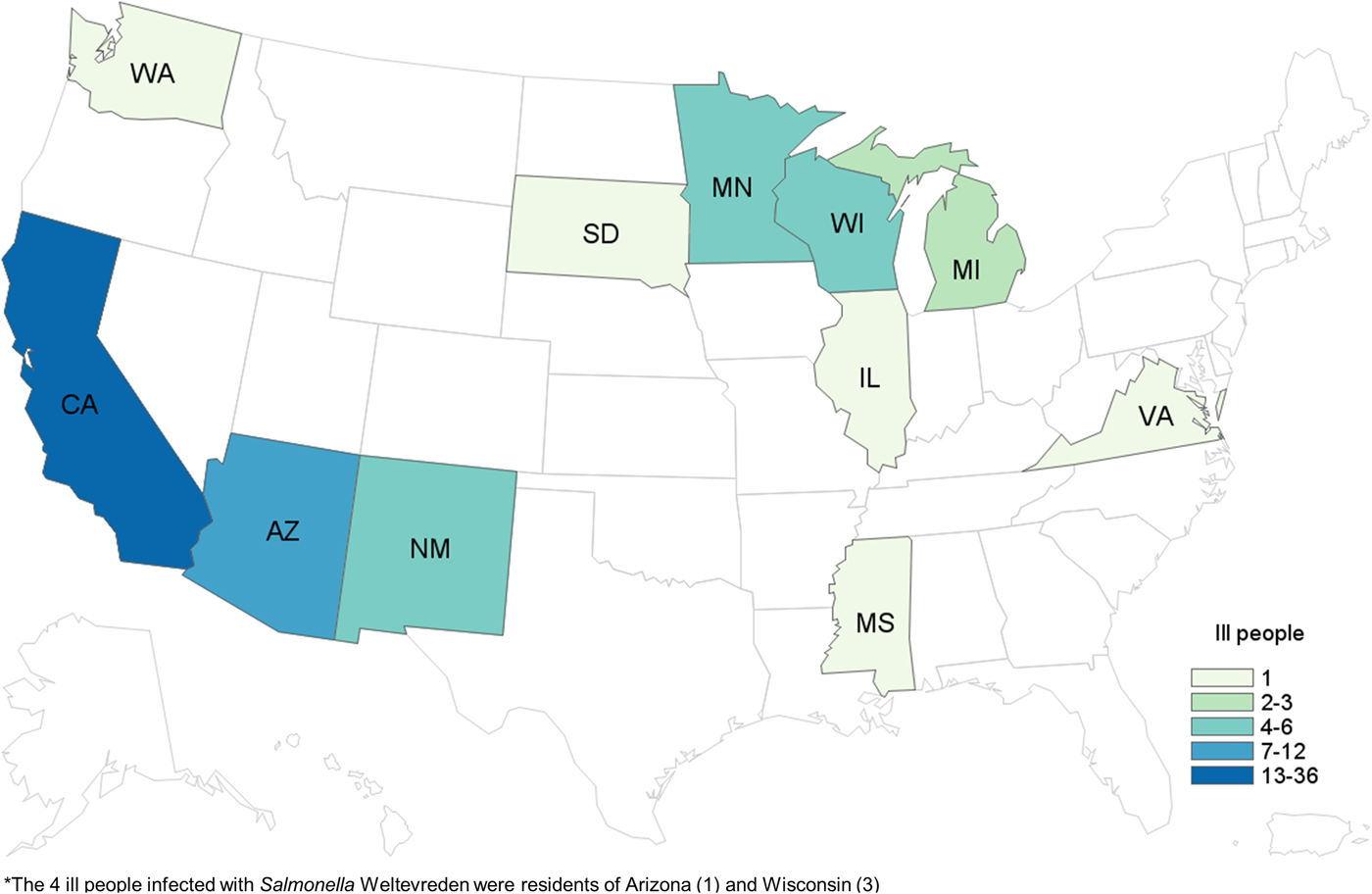
Fig. 1. People infected with the outbreak strains of Salmonella Paratyphi B var. L(+) tartrate(+) (n = 65) and Salmonella Weltevreden (n = 4), by state of residence*. *The four ill people infected with Salmonella Weltevreden were residents of Arizona (one) and Wisconsin (three).
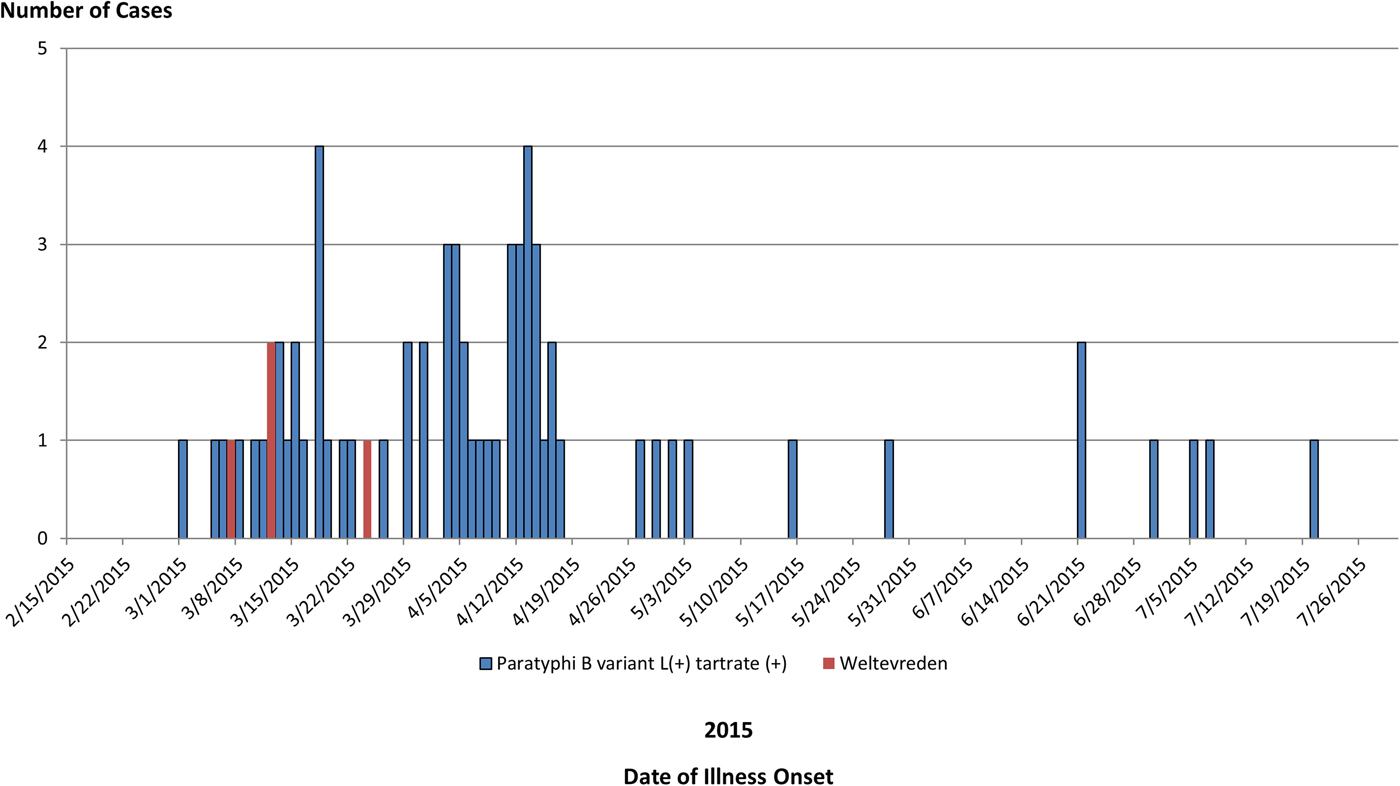
Fig. 2. People infected with the outbreak strains of Salmonella Paratyphi B var. L(+) tartrate(+) (n = 65) or Salmonella Weltevreden (n = 4), by date of illness onset.
Epidemiological investigation
By April 14, eight (57%) of 14 case-patients reported either eating seafood or dining at an Asian-style restaurant in California in the week before illness onset. Local and state investigators began interviewing case-patients with a more detailed hypothesis generating questionnaire, and 11 (92%) of 12 case-patients interviewed with this questionnaire reported eating sushi from a restaurant or grocery store in the week before illness onset. This was significantly higher than the 5% of healthy people who reported eating ‘sushi, sashimi or ceviche made with raw fish or shellfish’ in the 7 days before interview in the 2006–2007 FoodNet Population Survey (P < 0.05). All 11 case-patients reported consuming sushi containing raw tuna. However, eight of these 11 case-patients also reported consuming sushi containing other raw seafood, including salmon and yellowtail. On April 20, we deployed a focused questionnaire for use in the multistate investigation to gather detailed information about the sushi items consumed, including the specific ingredients, how they were prepared, sides and garnishes eaten with sushi and any other seafood items consumed. We identified five restaurant sub-clusters at sushi restaurants in Arizona (one), California (three) and New Mexico (one), each comprised of two to six case-patients. Detailed exposure information was available for three of the five sub-clusters in Arizona (one) and California (two). The only common seafood ingredient consumed by case-patients across all three sub-clusters was raw tuna. All case-patients in all three of these sub-clusters ate a sushi roll containing raw ‘spicy tuna’ (small diced or ground raw tuna mixed with a spicy sauce). In two of these three sub-clusters, all case-patients consumed multiple sushi items and had several seafood exposures other than spicy tuna in common, such as imitation crab or cooked shrimp. In the third sub-cluster, the only common seafood ingredient consumed among all five case-patients was spicy tuna.
By the end of the investigation, hypothesis-generating and/or focused interviews were conducted with 54 case-patients. Of 50 case-patients with available information, 46 (92%) reported consuming sushi in the week before they became ill. Of the 45 people reporting details of their sushi exposure, 44 (98%) reported eating a sushi item containing raw tuna, and 29 (88%) of 33 who specified type of tuna reported eating a sushi item containing raw spicy tuna.
Traceback investigation and product testing
Based on early interview information among case-patients in their state, California investigators began traceback investigations in April to determine if there was a common source for sushi ingredients reported by case-patients. As additional cases were reported in other states and as more epidemiological information became available, other state regulatory agencies and FDA conducted traceback to identify the sources of tuna used to make the tuna sushi consumed by case-patients. Ultimately, investigators selected seven points of service in four states as candidates for traceback, including four of the restaurant sub-clusters and three points of service reported by individual case-patients (Fig. 3). Investigators determined that these points of service used different recipes to prepare sushi rolls reported by case-patients, which included frozen raw ground, minced or chunk yellowfin tuna. Six of the seven points of service received frozen raw tuna from one common importer (importer B). Between December 2014 and March 2015, importer B received frozen raw tuna products from three Indonesian processors (processors B, C and D) that could have accounted for product served at these points of service.
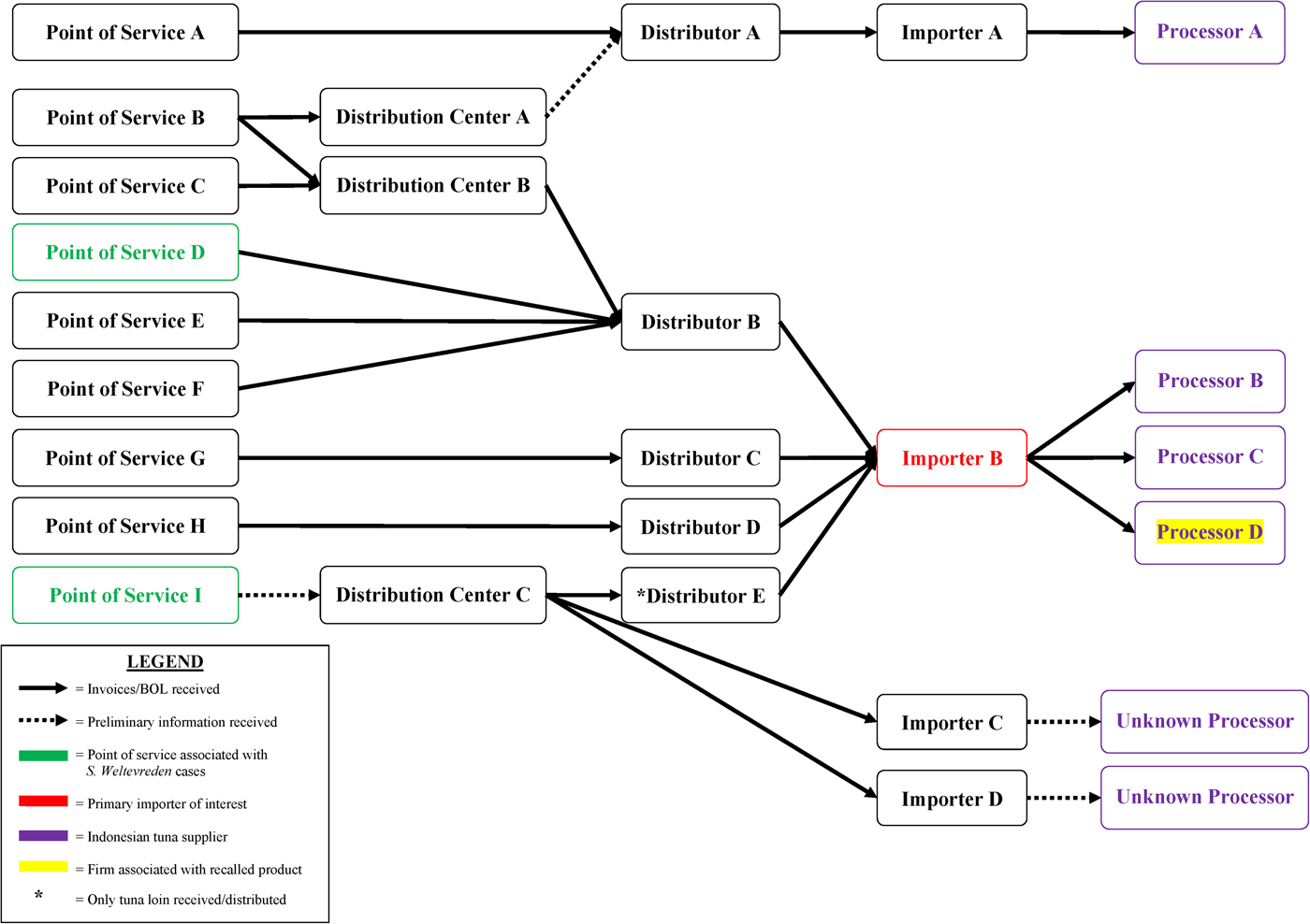
Fig. 3. Salmonella Paratyphi B variant L(+) tartrate(+) and Salmonella Weltevreden traceback investigation flow diagram for ground and chunk tuna. This diagram illustrates the flow of ground and chunk tuna from points of services where case-patients ate or bought raw tuna sushi to its origin.
From May through July 2015, a total of 87 samples of frozen raw tuna (saku, chunk, slice, strip, fillet and ground tuna) from nine distributors or points of service were collected by local, state and FDA investigators. Six of the samples were collected by FDA investigators and 81 samples were collected by local and state investigators in Arizona (15), California (46), Minnesota (15) and New Mexico (five). Salmonella was isolated from 12 (14%) of the 87 samples of frozen raw tuna; all 12 samples were supplied by processor D in Indonesia.
In May 2015, Arizona investigators collected samples of 15 unopened packages of frozen raw tuna for Salmonella testing: six samples of ground tuna from a restaurant identified from a consumer complaint, seven samples of ground tuna and one sample of sashimi tuna from distribution centre B which supplied the restaurant, and one bigeye tuna sample from an additional distributor not represented on the traceback diagram (Fig. 3). The consumer complaint contained a report of diarrhoeal illness, which was not confirmed as a case in this outbreak. Two of the 15 product samples yielded Salmonella, both of which were frozen raw ground tuna received by importer B, which sourced the tuna from processor D in Indonesia. One sample yielded a novel strain of Salmonella Newport and the other sample yielded a rare strain of Salmonella Weltevreden.
In July 2015, investigators in Minnesota collected 15 samples of unopened frozen raw chunk tuna from a grocery store where a case-patient reported buying spicy tuna sushi. These samples were from the same lot of product that was suspected to have been served to the case-patient and were sourced from processor D in Indonesia through importer B. Nine samples yielded Salmonella Paratyphi B (dT+) with the main outbreak PFGE pattern (JKXX01.1495). An additional sample yielded Salmonella Weltevreden with two different PFGE patterns; neither pattern was the same one isolated from the above-mentioned raw ground tuna sample from Arizona.
Outbreak-associated Salmonella Weltevreden cases
The investigation identified four case-patients infected with two different strains of Salmonella Weltevreden linked to this outbreak based on the information obtained from the epidemiological and traceback investigations.
Of the three Salmonella Weltevreden strains isolated from tuna samples collected in Arizona and Minnesota, two of these strains did not have any recent clinical matches in the PulseNet database: one strain was new to the database and one strain had no matching clinical isolates identified by PulseNet since 2011. PulseNet identified one clinical isolate matching the third Salmonella Weltevreden strain (PFGE XbaI pattern JQPX01.0371), which was isolated from an Arizona patient in March 2015. This Arizona case-patient became ill on 24 March 2015, and, upon interview with the outbreak-specific questionnaire, reported consuming a spicy tuna sushi roll at a California restaurant in the week before illness onset.
During the Salmonella Paratyphi B (dT+) investigation, Wisconsin investigators notified CDC of a local outbreak of Salmonella Weltevreden infections that they suspected was linked to raw tuna. This outbreak occurred in March 2015, and consisted of three people infected with a very rare strain of Salmonella Weltevreden (PFGE Xbal pattern JQPX01.0376). Illness onset dates ranged from 7 March 2015 to 12 March 2015. All three case-patients (100%) reported consuming sushi from the same local grocery store in the week before illness. One case-patient reported definitely consuming sushi with raw tuna, and the remaining two case-patients reported likely consuming a sushi roll with raw tuna.
FDA and state investigators conducted traceback investigations into the source of the raw tuna supplied to the points of service in California and Wisconsin. Traceback identified that the raw frozen tuna used at both locations to make sushi rolls reported by these case-patients was supplied by importer B and sourced from processor D in Indonesia, the same two firms implicated in the Salmonella Paratyphi B (dT+) illnesses (Fig. 3).
Laboratory investigation
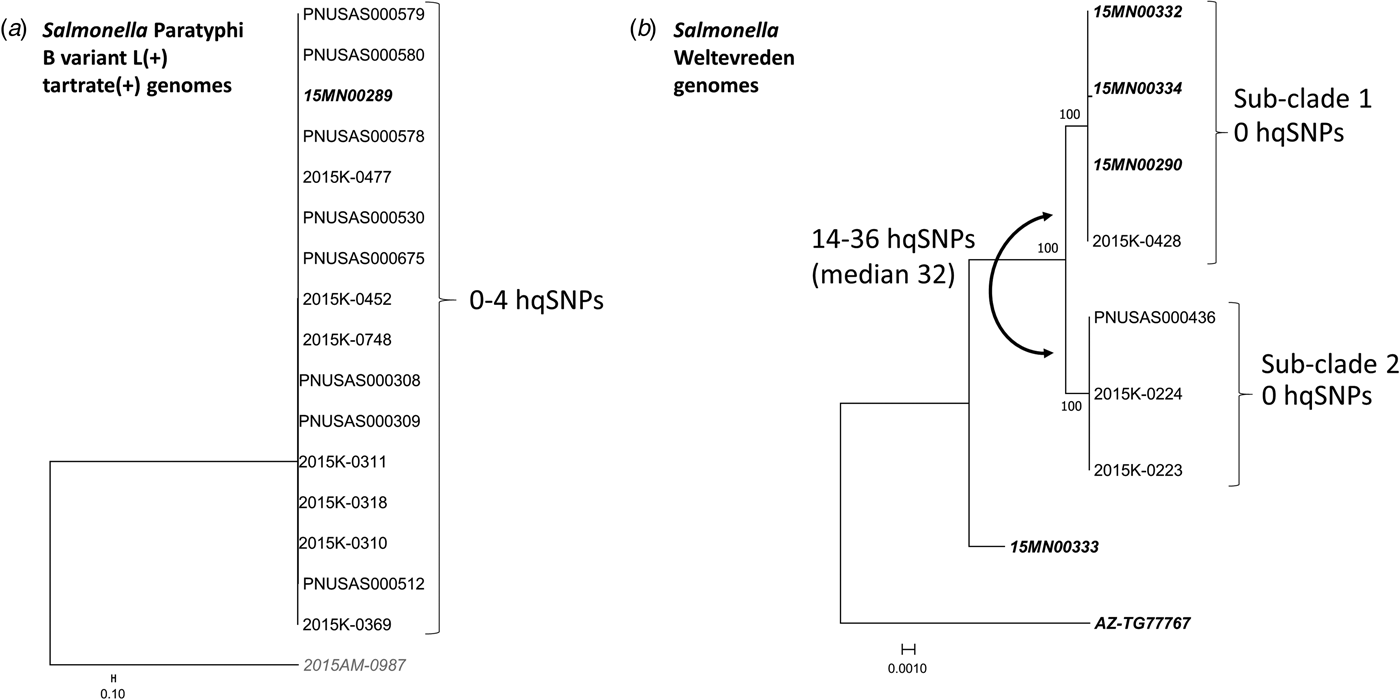
WGS was performed on 16 Salmonella Paratyphi B (dT+) isolates comprising both outbreak-associated PFGE patterns (Fig. 4a): 15 clinical isolates and one raw tuna isolate collected from a point of service in Minnesota were found to be closely related genetically (0–4 hqSNP differences out of a total of 879 informative positions) (Fig. 4a).

Fig. 4. Salmonella Paratyphi B variant L(+) tartrate(+) and Salmonella Weltevreden whole-genome sequencing analysis. Phylogenetic analysis of Salmonella Paratyphi B variant L(+) tartrate(+) (a) and Salmonella Weltevreden genomes (b). Genomes in bold italics are from tuna samples, remaining are clinical samples. The relevant SNP differences and noted next to brackets on the right of outbreak clades; differences between clades are noted to the left. (a) Sixteen Salmonella Paratyphi B (dT+) genomes (15 clinical and one tuna) comprising both outbreak-associated PFGE patterns are closely related genetically (0–4 nqSNPs). Genome 2015AM-0987 is a non-outbreak-associated outgroup. (b) Sub-clade 1 includes three tuna genomes from samples collected in Minnesota and one clinical genome from an Arizona case-patient; all are closely related (0 hqSNPs). Sub-clade 2 includes three clinical genomes from Wisconsin case-patients; all are closely related (0 hqSNPs). The two remaining genomes not included in either sub-clade are from tuna samples collected in Arizona and Minnesota; neither is closely related to other genomes in the analysis (70–450 hqSNPs, median 314 hqSNPs).
After PulseNet identified that the Salmonella Weltevreden strain isolated from one of the tuna samples collected in Minnesota shared an indistinguishable PFGE pattern with an Arizona clinical isolate from March 2015, both strains were analysed using WGS and were found to be closely related to each other (0 hqSNP differences out of a total of 535 informative positions). WGS was also performed on the three Salmonella Weltevreden clinical isolates from the local Wisconsin outbreak (PFGE Xbal pattern JQPX01.0376) and all were closely related genetically (0 hqSNP difference) to each other; however, these isolates were more distantly related to the two Salmonella Weltevreden from Minnesota and Arizona mentioned above (median difference of 32 hqSNPs; range 14–36 hqSNPs). WGS was performed on the two additional strains of Salmonella Weltevreden isolated from tuna tested in Arizona and Minnesota, and these isolates were genetically distinct from the other four Salmonella Weltevreden isolates described above (median difference of 314 hqSNPs; range 70–450 hqSNPs).
Control measures
On 21 May 2015, CDC and FDA publicly announced the multistate investigation indicating that consumption of sushi made with raw tuna from a yet to be determined common supplier was a possible source of the infections. As the investigation continued, importer B issued three voluntary recalls of frozen raw tuna imported from Indonesian processor D. First, on 27 May 2015, importer B recalled two lots of frozen raw ground tuna after being notified that Salmonella was isolated from samples of these lots collected in Arizona. On 20 July 2015, the importer announced a voluntary recall for one additional lot of frozen raw yellow fin tuna chunk meat sold to one distributor, after outbreak strains of Salmonella Paratyphi B (dT+) and Salmonella Weltevreden were isolated from samples of this lot tested in Minnesota. On 21 July 2015, importer B announced an expanded recall for all frozen raw tuna products (loin, saku, chunk, slice and ground market forms) sourced from processor D, which were sold from 9 May 2014 to 9 July 2015.
As a result of this investigation, FDA added processor D to an import alert, which allowed for the automatic detention of seafood products from this supplier due to the presence Salmonella [15]. Under this import alert, tuna shipments from processor D are to be denied entry into the USA until sufficient evidence is provided to the FDA demonstrating that the product is absent of Salmonella, and the import alert has been lifted for this firm.
Discussion
The combined epidemiological, traceback and laboratory evidence indicated that this national outbreak of Salmonella infections was associated with frozen raw ground and chunk tuna imported from Indonesia and consumed raw in sushi at restaurants and retail locations in the USA. This outbreak investigation highlights the continuing risk of foodborne illness associated with the consumption of raw seafood, such as raw tuna.
PFGE has been instrumental in foodborne outbreak detection over the past 20 years, particularly for dispersed multistate outbreaks. However, PFGE has limited discriminatory capabilities for some Salmonella serotypes, and similarities in PFGE banding patterns are not directly indicative of genetic relatedness. WGS is increasingly used during outbreak investigations, as it offers improved resolution to assess evolutionary relationships between isolates [Reference den Bakker16–Reference Jackson18]. While WGS is routinely being implemented for multistate listeriosis outbreaks [Reference Jackson18], this investigation is an example of the utility of WGS for a multistate Salmonella outbreak. In this outbreak, WGS provided additional microbiological evidence that diverse Salmonella Paratyphi B (dT+) PFGE patterns from clinical isolates were genetically related to each other and to tuna isolates. Additionally, we used WGS to microbiologically link a single Salmonella Weltevreden clinical isolate from Arizona to a tuna isolate from Minnesota.
The average annual number of foodborne outbreaks in the USA attributed to an imported food has increased from three annually during 1996–2000 to 18 annually during 2009–2014, and 55% of imported food outbreaks during this time frame were attributed to seafood [Reference Gould19]. This outbreak highlights the continued importance of food safety measures for imported seafood products. Specifically, this outbreak underscores the need for processors that import food into the USA to have and implement sanitation practices that prevent the contamination of seafood products with microbial pathogens, particularly in seafood products that are often consumed raw, such as in sushi.
Although an inspection at processor D was not conducted to assess possible root causes of contamination for this outbreak, many steps in tuna capture, harvesting and processing have the potential to cause contamination. Some examples of potential modes of contamination include contaminated water used to clean, cut or process tuna, ill food handlers contaminating tuna during harvest or processing, cross-contamination from processing equipment, or introduction during harvest or processing from animals, rodents or another source [Reference Iwamoto20]. A better understanding of tuna harvesting and processing practices in countries exporting raw tuna to the USA will help determine potential routes of bacterial contamination and help to prevent future outbreaks.
Finally, this investigation highlights the continuing risk of salmonellosis from eating raw seafood. This outbreak is the fourth large, multistate outbreak of Salmonella infections associated with raw tuna since 2007 [2]. Certain groups of people are at higher risk of acquiring a foodborne illness, such as children younger than 5 years or adults older than 65 years old, people with compromised immune systems and pregnant woman [21]. To reduce the risk of foodborne illness, these people should be especially careful to avoid undercooked animal products, such as raw seafood, at all times, not only during a known outbreak [22–23].
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0950268818001462.
Conflicts of interest
None.
Acknowledgements
The authors thank the following local, state and federal public health and regulatory officials and agencies for their contributions to the epidemiological, laboratory and environmental investigations during this outbreak: Arizona Department of Health Services, Maricopa County Department of Public Health, Maricopa County Environmental Services Department, California Department of Public Health, California Department of Public Health Microbial Disease Laboratory, County of San Diego Health and Human Services Agency, San Diego County Department of Environmental Health, Los Angeles County Department of Public Health, Orange County Health Care Agency, County of Riverside Department of Environmental Health, County of Riverside Department of Public Health, Ventura County Public Health, Ventura County Resource Management Agency, Contra Costa County Health Services, Illinois Department of Public Health, Michigan Department of Health and Human Services, Minnesota Department of Health, Minnesota Department of Agriculture Laboratory, Mississippi State Department of Health, New Mexico Department of Health, City of Albuquerque Environmental Health Department, South Dakota Department of Health, Virginia Department of Health, Washington State Department of Health, Wisconsin Department of Health Services, FDA Office of Regulatory Affairs, FDA Center for Food Safety and Applied Nutrition, FDA Office of the Commissioner.
Disclaimer
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.