Introduction
Enteric illnesses cause considerable morbidity and mortality worldwide. Waterborne enteric diseases cause 2 million deaths each year, the majority of which occur in children aged 5 and under [1]. Foodborne enteric diseases are responsible for 600 million illnesses and 420 000 deaths annually [2]. These illnesses impact the quality of life of those affected and result in enormous financial consequences for individuals and nations [Reference McLinden3]. Although most enteric illnesses are transient, significant chronic sequelae associated with some foodborne pathogens can have long-term public health impacts [Reference Keithlin4–Reference Scallan6].
Enteric illness outbreak investigations seek to identify the source of illnesses to prevent further illness in the population. Timely source identification is a key step towards reducing the incidence of enteric illness worldwide and can lead to change in public health policy or recommendations to prevent future outbreaks, such as changes to food manufacturing processes or regulations. Timely source identification can also lead to public health notices and recalls that may prevent further illnesses in a specific outbreak. Accurate source identification can also provide opportunities to learn more about known and emerging diseases, increase understanding of the impact of current disease prevention practices and improve public confidence in public health agencies responsiveness to disease outbreaks [Reference Reingold7].
Outbreak investigations take many forms, depending on the pathogen, context, affected population and suspected route of transmission. Initial cases often alert public health officials that a possible outbreak is occurring. Once an outbreak has been identified a case definition is established to support case finding activities. As cases are identified, information is gathered about the outbreak to generate hypotheses about the potential source(s) and route(s) of exposure. Information can come from a range of sources, including the cases themselves, their friends or family, staff members of businesses and institutions, experts or literature and physical and environmental sampling and inspections. Taken together, this information supports the development of hypotheses about the source of the outbreak.
Hypothesis generation about both the potential source(s) and route(s) of exposure is a key step in outbreak investigations, as it begins the process of narrowing the search for the transmission vehicle. Although some hypothesis generation methods have been described in summaries of outbreak investigation steps [Reference Reingold7, 8], the full range of possible methods used in outbreak investigations or the frequency that they are used is not readily available. We conducted a scoping review to summarise the methods for hypothesis generation used during human enteric illness investigations and to understand the frequency and breadth of methods, as well as to identify knowledge gaps and areas for future research.
Methods
A scoping review protocol was created a priori using the framework established by Arksey and O'Malley [Reference Arksey and O'Malley9]. A copy of the protocol, including the search strategy, the screening tool and the data characterisation tool can be found in Supplementary Material S1. A full list of the articles identified in this scoping review can be found in Supplementary Material S2. A review team was established and included expertise in synthesis research, food safety, epidemiology and outbreak investigation.
The research question:
What methods have been used, or could be used, in human enteric illness outbreak investigations for hypothesis generation?
Search terms and strategy
A search algorithm (Supplementary Material S1) was constructed using key terms from 30 pre-selected relevant articles and implemented in five databases (PubMed, Scopus, Embase, Cumulative Index to Nursing and Allied Health Literature (CINAHL) and ProQuest Public Health) on 25 May 2015 with a date filter of 1 January 2000–25 May 2015.
The search was evaluated for capture sensitivity by searching reference lists of 12 randomly selected relevant primary methodology papers and 10 of the most recent relevant literature reviews in PubMed (Supplementary Material S1). The grey literature search targeted websites of government and research organisations, and relevant articles from Conference Proceedings (Supplementary Material S1). A total of 202 articles were identified by the grey literature search that were not captured by the search strategy and were added to the literature review (Fig. 1). All citations were exported and de-duplicated in RefWorks (ProQuest, LLC), an online bibliographic management program, before being uploaded into a web-based systematic review management program, DistillerSR™ (Evidence Partners, Ottawa, Canada), for evaluation and characterisation.
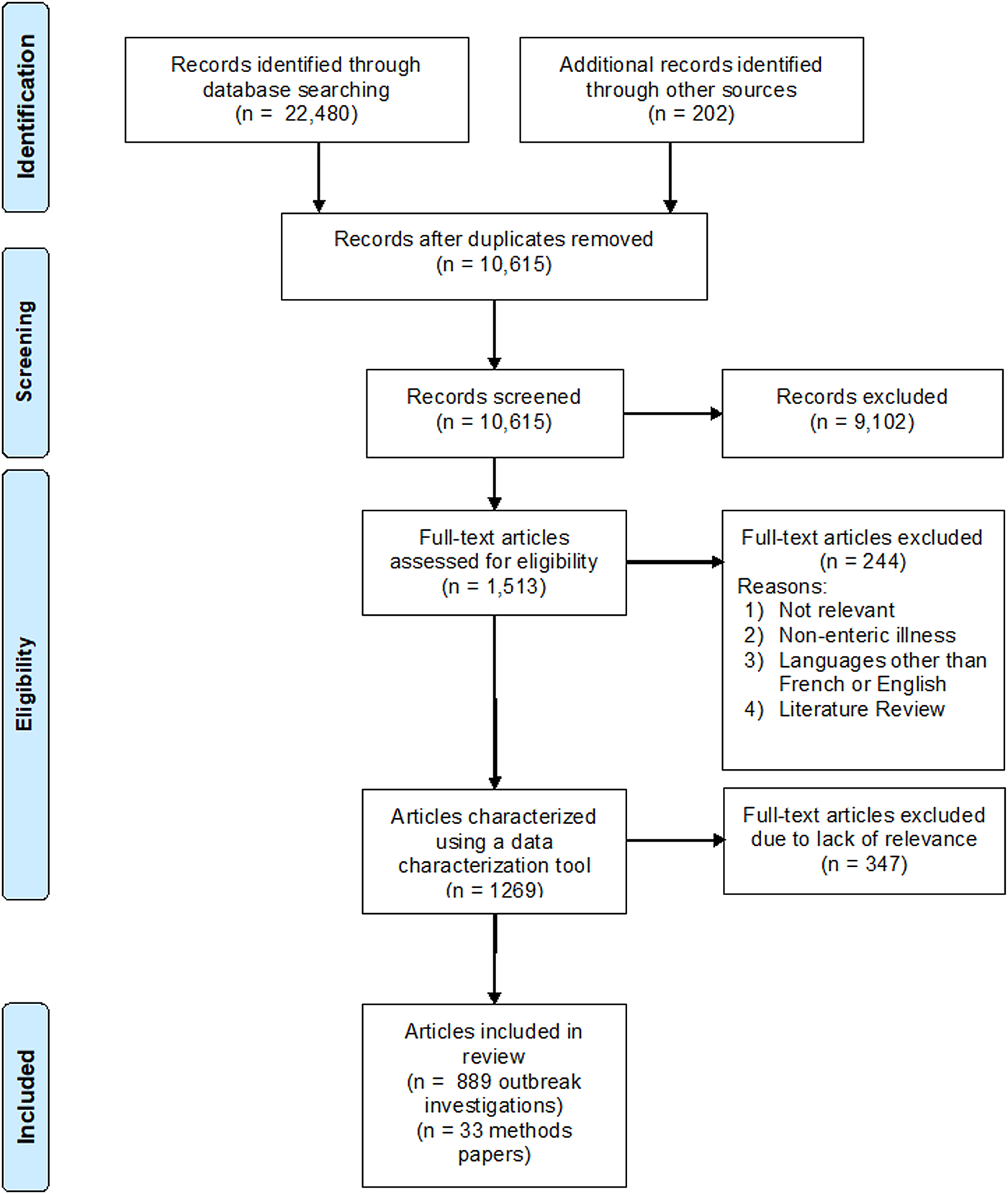
Fig. 1. PRISMA flow chart documenting the literature retrieval and inclusion/exclusion criteria for citations to identify methods of hypothesis generation during human illness investigations.
Relevance screening of abstracts and full-text citation
Each title and abstract was screened by two independent reviewers using a relevance screening form (Supplementary Material S1). Articles were included if they met the following criteria: (1) used or described methods applicable to enteric illness outbreak investigations to assist in hypothesis generation and source identification; (2) published after 1 January 2000 and (3) were reported in either English or French language. No geographic location was used as an exclusion criterion. The relevance screening form was pretested on 50 citations and resulted in a kappa agreement >0.8, indicating good agreement. Two reviewers screened each citation independently and conflicts were resolved by consensus.
Potentially relevant articles were procured, confirmed to be in English or French and relevant before broadly being characterised by two independent reviewers using a secondary relevance screening tool (Supplementary Material S1) to gather information on the outbreak, such as geographic location, type of pathogen, setting (single or general) and implicated source (Supplementary Material S1). This form was pretested on 10 papers to ensure good agreement and clarity within the form.
Data extraction and analysis
The data characterisation and utility tool was used to gather data on the hypothesis generation methods used in the outbreak investigation. The form contained check boxes for 23 known hypothesis generation methods and an option for reviewers to add other methods not captured in the form. Clearly established definitions were used to help data extractors distinguish between instances when a method was used for hypothesis generation or hypothesis testing. Hypothesis generation was defined as the process of developing one or more tentative explanations about the source of the outbreak used to inform further investigation. This was distinguished from hypothesis testing, which was defined as the process of confirming that a specific exposure is or is not the cause of an outbreak. Hypothesis testing is performed on a small number of suspect exposures and may include statistical testing or traceback investigation. Sometimes, when the hypothesis is refuted, additional rounds of hypothesis generation may be initiated. Several methods included in the form could be used for either hypothesis generation or for hypothesis testing in outbreak investigations. For example, analytic studies can be used to examine a wide range of exposures to help generate hypotheses about plausible sources. However, analytic studies can also be used to test a hypothesis when a specific source is suspected. Instances where methods were used to test a hypothesis were not relevant to this review and were not captured on the form. Where more than one outbreak was described in a single paper, multiple forms were completed to capture methods used in different investigations. This form was pretested on five papers to ensure agreement between reviewers was adequate and to improve the clarity of the questions/answers where necessary. Two reviewers independently reviewed each paper and disagreements between reviewers were discussed until a consensus was reached or settled with a third reviewer. Articles with no hypothesis generation methods described or with a known source at the outset of the investigation were excluded at this stage. Papers describing methodology, but not specific outbreak investigations, were identified and are described separately. Descriptive statistics were used to summarise the dataset using Stata 15 (StataCorp, 2017).
Results
In total, there were 10 615 unique citations captured by the search (Fig. 1). Of these, 889 (8.4%) papers were fully characterised and included 903 reported outbreaks (Supplementary Material S2). Of the reported outbreaks, 25 (2.8%) were described in 11 multi-outbreak articles and the remaining 878 (97.2%) were described in single outbreak articles (Fig. 1).
The pathogens associated with the outbreaks included: bacteria (n = 622, 68.9%), viruses (n = 192, 21.3%), parasites (n = 64, 7.1%), bio-toxins (n = 3, 0.3%), fungi (n = 1, 0.1%) and multiple pathogens (n = 11, 1.2%). The pathogen was not identified in 10 (1.1%) outbreaks. In terms of outbreak source, 552 (61.1%) identified food as the source, while 103 (11.4%) identified water, 34 (3.8%) identified direct contact with animals, 25 (2.8%) identified person-to-person transmission, 25 (2.8%) identified multiple modes of transmission, 20 (2.2%) identified food-handlers, 8 (0.9%) identified soil or environment and 5 (0.6%) reported other modes of transmission as the source. In 131 (14.5%) of the outbreaks, no source was identified.
Hypothesis generation methods used in the enteric illness outbreak investigations are listed and defined in Table 1. The majority (n = 733, 81.2%) of investigations employed two or more methods to generate hypotheses; the median number of methods used was three (interquartile range: 2–4). Analytic studies (n = 585, 64.8%) were the most commonly reported method category, followed by descriptive epidemiology (n = 304, 33.7%), and food or environmental sampling (n = 296, 32.8%). Uncommon methods included tracer testing (n = 1, 0.1%), anthropological investigation (n = 1, 0.1%) and industry consultation (n = 1, 0.1%).
Table 1. Description and frequency of methods used to generate a hypothesis in 903 human enteric illness outbreak investigations identified in scoping review citations
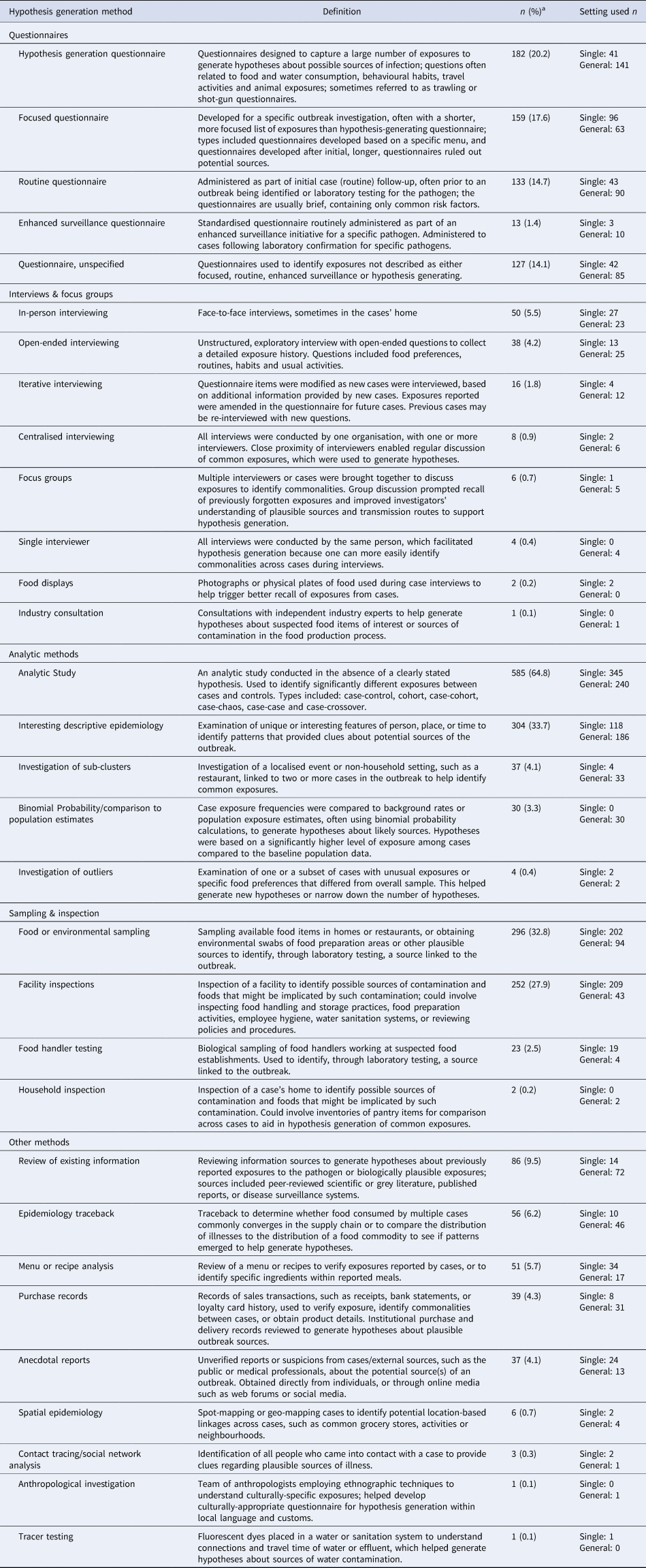
a Percentages will not sum to 100% as outbreak investigators could use multiple methods to generate hypotheses.
Single setting outbreaks
The proportion that each method was used within single setting outbreaks, such as a restaurant, nursing home, or event, is reported in Figure 2. The most commonly reported methods used in single setting outbreaks included analytic studies (n = 345, 27.2%), facility inspections (n = 209, 16.5%) and food or environmental sampling (n = 202, 15.9%). The least common methods used in single setting outbreaks included focus groups (n = 1, 0.1%) and tracer testing (n = 1, 0.1%). Binomial probability/comparison to population estimates, single interviewer and anthropological investigation were not reported in single setting outbreaks.
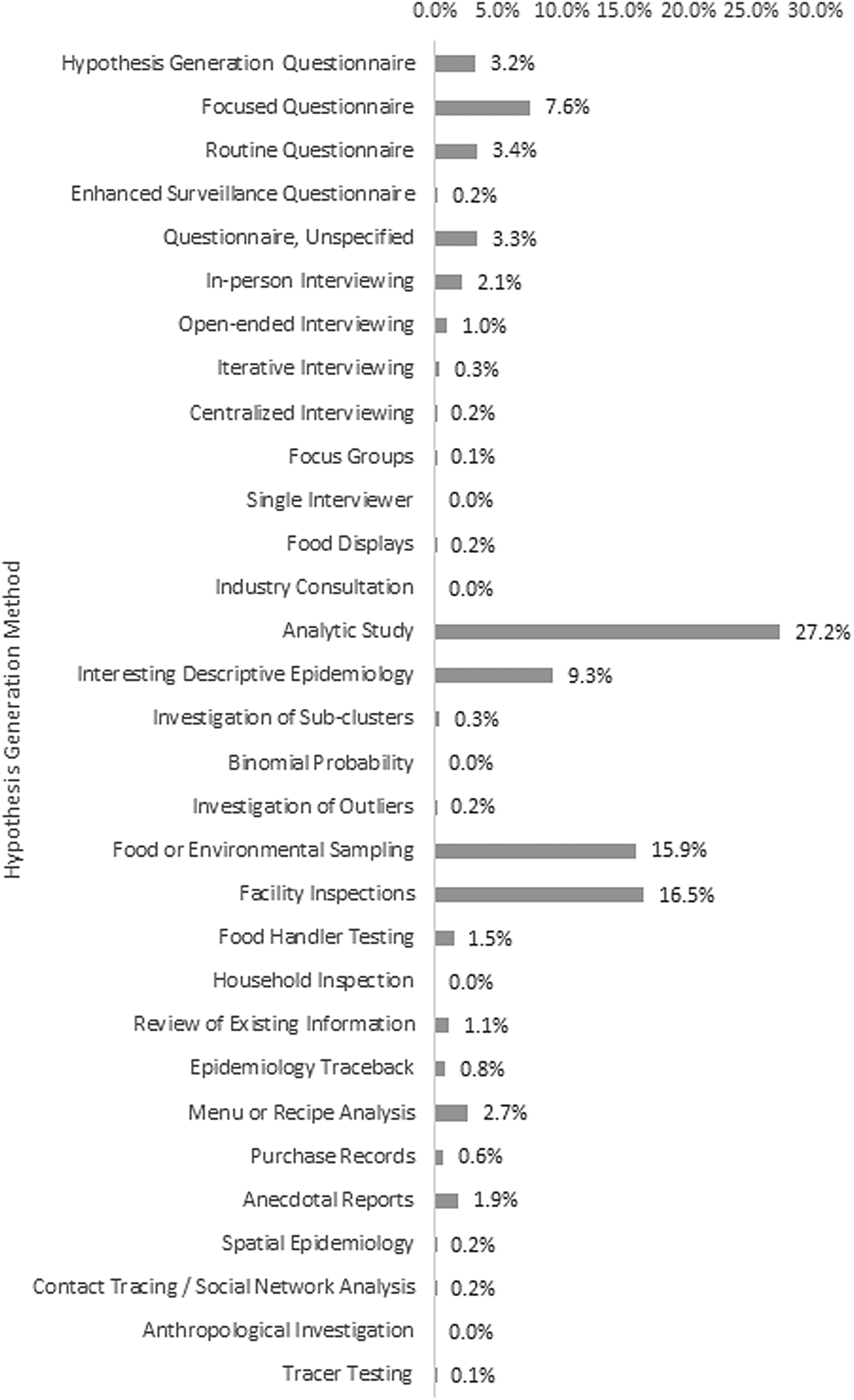
Fig. 2. Hypothesis generation methods used in single setting outbreaks.
General population outbreaks
The proportion that each method was used in general population outbreaks, outbreaks not related to a single event or venue, is reported in Figure 3. The most commonly used methods in general population outbreaks included analytic studies (n = 240, 18.7%), interesting descriptive epidemiology (n = 186, 14.5%) and hypothesis generation questionnaires (n = 141, 11.0%). The least common methods used in general population outbreaks included anthropological investigation (n = 1, 0.1%), contact tracing/social network analysis (n = 1, 0.1%) and industry consultation (n = 1, 0.1%). Tracer testing and food displays were not reported in general population outbreaks.

Fig. 3. Hypothesis generation methods used in general population outbreaks.
Hypothesis generation innovation and trends 2000–2015
Trends in method use over the 15-year span were examined in 5-year increments (Supplementary Material S3). Small increases were observed in the use of anecdotal reports, purchase records, binomial probability/population comparison, facility inspections and review of existing information. A decline was observed in the use of analytic studies. Other methods had variable use over the time period or were relatively stable.
Methodology papers
Of the 10 615 citations screened, 33 (0.3%) methods papers were identified (Supplementary Material S2). These papers focused on evaluating existing methods or comparing standard vs. a novel approach to hypothesis generation (Supplementary Material S4). Of these, the most commonly discussed method was analytic studies (n = 11, 33.3%). This included five on the validity of case-chaos methodology [Reference Doerken10–Reference Edelstein, Wallensten and Kuhlmann-Berenzon14], two on case-case methodology [Reference Gillespie15, Reference Wilson16], two on case-control methodology [Reference Gu17, Reference Jervis18], one discussing the validity of case-cohort methodology [Reference de Waroux, Maguire and Moren19] and one discussing the validity of case-crossover methodology [Reference Haegebaert, Duche and Desenclos20].
The use of laboratory methods, including whole genome sequencing, was described in five (15.2%) papers [Reference Gilpin21–Reference De Lappe and Cormican25]. Traceback procedures were explored in five (15.2%) papers, including three on the use of network analysis [Reference Doerr26–Reference Yan28], one on the use of food flow information [Reference Huber and Luber29] and one examining the use of relational systems to identify sources common to different cases [Reference Weiser30]. Four (12.1%) papers described broad outbreak investigation activities, which included the hypothesis generation step, one from the United Kingdom [Reference Harker31], one from Quebec, Canada [Reference Gaulin32], one from Minnesota [Reference Rounds33] and one from the Centers for Disease Control and Prevention (CDC) in the United States [Reference Sobel34]. Three (9.1%) papers explored interviewing techniques, two examining the use of computer assisted telephone interviews (CATI) technology [Reference Fox35, Reference Kirk36] and one on when to collect interview-intensive dose-response data [Reference Jones37]. Three (9.1%) papers compared online questionnaires to phone or paper questionnaires [Reference Oh38–Reference Rosner and Morrison40]. Finally, one (3.0%) paper examined the use of mathematical topology methods to generate hypotheses [Reference Buscema41] and another (3.0%) paper examined the use of sales record data to generate hypotheses [Reference Kaufman42].
Discussion
The most commonly reported hypothesis generation methods identified in this scoping review included analytic studies, descriptive epidemiology, food or environmental sampling and facility inspections. Uncommon methods included industry consultation, tracer testing, anthropologic investigations and the use of food displays. Most outbreak investigations employed multiple methods to generate hypotheses and the context of the outbreak was an important determinant for some methods.
The multitude of hypothesis generation methods described and the use of multiple methods by most outbreak investigators point to the complexity of investigating enteric illness outbreaks. Many methods described are complementary with other methods or may be used in sequence as an investigation progress. For example, routine and enhanced surveillance questionnaires will often be collected before an outbreak is even identified, while hypothesis generating questionnaires are frequently used at the beginning of an outbreak when the focus of the investigation is quite broad. The use of descriptive epidemiology is generally based on questionnaire data and is often one of the first hypothesis generation methods employed in outbreak investigations. Other methods, such as food or environmental sampling, facility inspections and food handler testing may be used in conjunction with questionnaires, particularly if the outbreak occurred in one setting or at an event. Both open-ended and iterative interviewing frequently occur later in investigations when no obvious source has emerged or as new cases are identified.
Investigators consider many factors when choosing a hypothesis generation method. For example, the length of time that has elapsed between case exposure and the identification of outbreak impact investigation tools such as the collection of contaminated food and environmental samples or facility inspections and traceback investigations [Reference Gutiérrez Garitano43–Reference Lienemann45]. Cost and feasibility are also important considerations for many hypothesis generation methods. Analytic studies can be expensive and time consuming [Reference Schmid46], while food and environmental sampling requires laboratory resources for testing [Reference Rakesh47, Reference Lee, Ong and Auw48]. Changes in method type used over time, for example increases in the use of anecdotal reports and purchase records, likely reflect the increase in available technology such as online reporting through social media, and availability of online records. The decline in the use of analytic methods may reflect the increased availability of other, less expensive, hypothesis generation methods such as population comparisons or purchase records.
Outbreak setting can impact the choice of hypothesis generation methods. Methods frequently used in single setting outbreaks include tailored menu-based interviewing, facility inspections and food handler testing. These methods are well-suited to these settings because the common connection across cases is obvious and the source is expected to be identified at a single location common to the cases, such as a restaurant or hospital. For outbreaks related to a single event such as weddings or conferences, analytic studies such as a retrospective cohort are well suited to investigating known exposed populations. In contrast, the use of purchase records, such as store loyalty cards or credit card statements, is utilised when the outbreak is among the general population and there appears to be no obvious connection between cases. Similarly, a review of existing information is a method used frequently in outbreaks among the general population when the range of plausible sources of illness is substantially larger than would be present in single event outbreaks. Outbreak setting thus has implications for the feasibility and usefulness of many hypothesis generation methods.
One finding of this scoping review is that hypothesis generation methods are not well reported within outbreak reports. Descriptions of hypothesis generation methods and sequence of events were often limited or entirely omitted from the publications. This incomplete reporting makes it difficult to interpret how frequently some methods are used by outbreak investigation teams compared to what outbreaks are written up and published in detail. Thus, it is likely that some common methods such as routine questionnaires were underreported and are thus underrepresented in this review. Methods that did not contribute to the identification of the source may also not be reported. Thorough reporting of all hypothesis generation methods used by outbreak investigators would allow for a more comprehensive understanding of the range and frequency of methods used to investigate outbreaks.
Most of the methods papers identified in this review focused on analytic studies, laboratory methods, traceback, interviews and questionnaires. No methods papers were identified related to several hypothesis generation methods reported in this review, including focus groups, iterative interviewing, open-ended interviewing, descriptive epidemiology, sub-cluster and outlier investigation, food or environmental sampling, facility inspections, food handler testing, review of existing information, menu or recipe analysis, anecdotal reports and social network analysis. The paucity of methods papers exploring hypothesis generation methods is an important literature gap. The relative merits of different hypothesis generation methods, their validity and reliability and comparable effectiveness across outbreak investigations, are needed to support outbreak investigator decision-making.
The frequencies of hypothesis generation methods reported in this scoping review may differ from their frequencies in practice as most outbreaks identified had successfully identified the source of the outbreak. Only 15% failed to identify the source of the outbreak, which is a much lower proportion than expected in practice [Reference Murphree49, 50]. This suggests that investigations where the source is not identified are less likely to be published and/or are published with few details, so they did not fulfil the inclusion criteria. This underreporting makes it impossible to accurately assess individual hypothesis generation methods' relative impact on investigation success based solely on published literature. Increased reporting of outbreak investigations where the source is not identified would improve our understanding of effective vs. ineffective hypothesis generation method use. Alternatively, organisations with access to administrative data on a full complement of outbreaks could analyse the relationship between the hypothesis generation methods used and associated outcomes of all outbreak investigations. For instance, Murphree et al. [Reference Murphree49] compared the success of analytic studies to other methods in identifying a food vehicle across all outbreaks in the United States Foodborne Diseases Active Surveillance Network (FoodNet) catchment area. Analytic studies had a 47% success rate compared to all other methods with a 14% success rate [Reference Murphree49], suggesting that analytic studies, where feasible, are more likely to lead to the identification of the source. However, given that analytic studies are not always feasible or appropriate, additional information on the relative success of other methods would help outbreak investigators choose appropriate methods to optimise the likelihood of successfully identifying the source. It would be valuable if outbreak investigators reported brief evaluations of their hypothesis generation methods to improve our understanding of the strengths and limitations of each method.
This review employed a comprehensive search strategy to identify enteric outbreak investigations and articles on hypothesis generation methods for outbreaks or other foodborne illness investigations. It is possible that despite our efforts some outbreak reports with hypothesis generation information were missed, as outbreaks are often not reported in the peer-reviewed literature and thus are not indexed in searchable bibliographic databases. To circumvent this shortfall, we performed a comprehensive grey literature search, however, it is possible some relevant reports were missed. It is also possible that there is some language bias, as the search was conducted in English and only papers reported in English or French were included in the review. This may have resulted in a failure of the search to identify relevant non-English papers. The effect of this on our results and conclusions is unknown. Lastly, because some methods identified in this review could be used for either hypothesis generation or hypothesis testing, we may have misclassified some uses of those methods as hypothesis generation when the investigators actually used the method for hypothesis testing. We relied on author reporting to understand when hypothesis generation was taking place, but incomplete or inadequate reporting may have resulted in misclassification that overestimated the extent to which some methods, such as analytic studies, are used to generate hypotheses.
This review demonstrated the range of hypothesis generation methods used in enteric illness outbreak investigations in humans. Most outbreaks were investigated using a combination of methods, highlighting the complexity of outbreak investigations and the requirement to have a suite of hypothesis generation approaches to choose from, as a single approach may not be appropriate in all situations. Research is needed to comprehensively understand the effectiveness of each hypothesis generation method in identifying the source of the outbreak, improving investigators' ability to choose the most suitable hypothesis generation methods to enable successful source identification.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0950268819001699.
Acknowledgements
The Public Health Agency of Canada library for their help in the procurement of publications. The Public Health Agency of Canada Centre for Food-borne, Environmental and Zoonotic Infectious Diseases, Outbreak Management Division contributors: Jennifer Cutler, Kristyn Franklin, Ashley Kerr, Vanessa Morton, Florence Tanguay, Joanne Tataryn, Kashmeera Meghnath, Mihaela Gheorghe, Shiona Glass-Kaastra.
Financial support
This research received no specific grant from any funding agency, commercial or not-for-profit sectors.
Conflict of interest
None.








