INTRODUCTION
Clinical and public health microbiology laboratories worldwide constitute a rich, underutilized resource in monitoring the changing epidemiology of microbial populations and in identifying emerging public health threats. Through the routine capture and interpretation of diagnostic test results such as organism identification and antimicrobial resistance phenotypes, electronic laboratory-based surveillance can improve the diagnostic specificity and response time of traditional infectious disease surveillance [Reference Leal and Laupland1–Reference Wurtz and Cameron3]. The ability to promptly detect outbreaks caused by highly resistant pathogens is of particular importance.
To realize the full potential of microbiology laboratory data for disease outbreak detection on a national scale, new tools and strategies for data management, analysis, interpretation, and communication are required. Visual monitoring of weekly case counts for pre-defined geographic units is inadequate for tracking hundreds of microbial species or thousands of phenotypically distinct pathogenic strains which do not respect artificial geographic boundaries. Time-trend analysis and other temporal statistics have been applied to laboratory data for the detection of public health outbreaks in the community [Reference Hutwagner4, Reference Widdowson5] and hospital setting [Reference Brown6, Reference Hacek7], but these do not address the geographic component of pathogen emergence and spread. Incorporating both time and geography, the prospective space–time permutation scan statistic [Reference Kulldorff8], which applies minimal assumptions concerning the time, length, and geographical size of potential outbreaks, is increasingly used for the early detection of disease outbreaks of public health importance [Reference Heffernan9–Reference Mostashari11].
The adoption of a uniform microbiology database by laboratories across Argentina through the Collaborative Group WHONET-Argentina provides a unique opportunity to apply such outbreak detection methods to laboratory data on a national scale and evaluate their performance. We selected Shigella sp. as a pathogen for an initial evaluation of the approach because of the public health importance of shigellosis outbreaks in Argentina and the particular challenge of detecting outbreaks in an organism which exhibits strong seasonal dependency. The early detection of outbreaks caused by foodborne and waterborne pathogens, such as Shigella, is key to tracing sources of contamination and implementing control measures. Because of the increased morbidity and mortality associated with multidrug-resistant Shigella infections [Reference Niyogi12, Reference Niyogi13], recent reports documenting the first appearance of highly resistant Shigella clones in Argentina are a cause for concern [Reference Andres14–Reference Rapoport16].
Our goal was to develop and assess the performance of WHONET and SaTScan for the detection of outbreaks of antimicrobial-resistant Shigella in Argentina as a first evaluation of the potential utility of such a system for other pathogens and locations. We were also interested in assessing the role of antimicrobial resistance phenotypes as a practical and specific marker for early detection of outbreak clones. This work was conducted under the National Institutes of Health Models of Infectious Disease Agent Study (MIDAS), a consortium whose goal is to develop and apply modelling methods for understanding the spread and detection of infectious agents.
METHODS
WHONET-Argentina laboratory data
The Collaborative Group WHONET-Argentina was established in 1986 under the coordination of the Antimicrobials Service of the Instituto Nacional de Enfermedades Infecciosas ANLIS ‘Dr C. Malbrán’ (INEI) to support laboratory training, epidemiological and molecular research, and public health surveillance of antimicrobial resistance [Reference Rossi17]. The network comprises 70 microbiology laboratories representing the 23 provinces of Argentina as well as the federal capital of Buenos Aires. Isolate-level data with available patient demographics and sample details are entered into WHONET, a free software developed by our group at the WHO Collaborating Centre for Surveillance of Antimicrobial Resistance and distributed through the World Health Organization to support antimicrobial resistance surveillance in more than 90 countries involving over 1000 clinical, public health, veterinary, and food microbiology laboratories [18, Reference Stelling and O'Brien19].
Participation in WHONET-Argentina is contingent upon ongoing acceptable performance in semi-annual proficiency tests of species identification and antimicrobial susceptibility testing. A total of 38 of the 70 WHONET-Argentina laboratories (Fig. 1) were included in this study based on compliance with national protocols for susceptibility testing, capacity for identification of Shigella isolates, and completeness of data entry for the period July 2005–June 2007. Data from 29 laboratories were available for the whole period, and data from an additional nine were available from 1 January 2006 onward. The primary reasons for excluding laboratories were insufficient data because of incomplete data entry (14 laboratories) or enrolment in the WHONET-Argentina network after January 2006 (15 laboratories). Three laboratories were excluded because Shigella isolates were usually identified only to genus level.
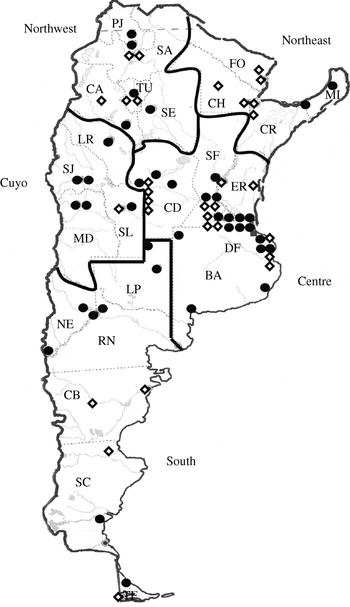
Fig. 1. Geographic distribution of laboratories in the Collaborative Group WHONET-Argentina. The 38 laboratories included in the analyses of this paper are indicated with a solid circle (•), while others are indicated with an open diamond (⋄). BA, Buenos Aires (province), CA, Catamarca, CB, Chubut, CD, Córdoba, CH, Chaco, CR, Corrientes, DF, Buenos Aires (federal capital), ER, Entre Ríos, FO, Formosa, LP, La Pampa, LR, La Rioja, MD, Mendoza, MI, Misiones, NE, Neuquén, PJ, Jujuy, RN, Río Negro, SA, Salta, SC, Santa Cruz, SE, Santiago del Estero, SF, Santa Fe, SJ, San Juan, SL, San Luis, TF, Tierra del Fuego, TU, Tucumán.
Antimicrobial susceptibility testing was performed by disk diffusion or broth microdilution in accordance with guidelines of the Clinical and Laboratory Standards Institute (CLSI) [20]. CLSI interpretive criteria were used for all antimicrobials with the exception of fosfomycin, for which recommendations of the French Society of Microbiology were used [21]. Susceptibility to six antimicrobials was tested by participating laboratories: ampicillin (AMP), ciprofloxacin (CIP), fosfomycin (FOS), nalidixic acid (NAL), nitrofurantoin (NIT), and trimethoprim/sulfamethoxazole (SXT). Resistance was rare to four of these agents (CIP, FOS, NAL, NIT), so we restricted analyses of resistance phenotypes to the two agents to which susceptibility was variable (AMP, SXT).
We looked for outbreaks at three levels of organism identification: (1) genus – Shigella; (2) species – S. flexneri, S. sonnei, S. dysenteriae, and S. boydii; and (3) resistance phenotype within species: non-susceptible to AMP alone (AMP), non-susceptible to SXT alone (SXT), non-susceptible to both agents (AMP–SXT), and susceptible to both (‘Susceptible’). Resistant and intermediate isolates were categorized as ‘non-susceptible’ for purposes of these analyses. Insufficient data were available to perform comparable analyses using Shigella serotype.
Through the National Diarrhoea and Foodborne Pathogens Laboratory Network, the Enteric Pathogens Service at INEI receives isolates of Shigella spp. collected during outbreaks as well as a sample of sporadic strains for serotyping. A subset is further subtyped by pulsed-field gel electrophoresis (PFGE) using standardized protocols from the PulseNet International Network to establish the genetic relatedness of clones and to confirm the occurrence of suspected outbreaks [Reference Ribot22, Reference Swaminathan23].
Space–time permutation scan statistic
The prospective detection of disease outbreaks poses specific challenges in that we know neither where nor when an outbreak may occur, nor its temporal or geographical extent. For the proposed surveillance system, we used a space–time permutation scan statistic [Reference Kulldorff8] as implemented in the free SaTScan™ software [24]. The method automatically adjusts for purely geographic patterns constant over time such as population density and for purely temporal trends nationwide such as day-of-week effects or seasonal patterns of disease occurrence. This automatic adjustment for seasonal variation – through the comparison of simultaneous temporal trends over multiple geographic units – offers a significant advantage over traditional purely temporal approaches which either do not adjust for seasonal trends [Reference Brown6, Reference Hutwagner25] or which require several years of comparable baseline data [Reference Hutwagner4, Reference Farrington26, Reference Farrington and Beale27].
The statistical likelihood that observed signals are due to chance alone is expressed in terms of a recurrence interval [Reference Kleinman28], which is the inverse of the P value. A signal with a recurrence interval of 180 days is of a strength that one would expect to see by chance alone about twice a year and thus consistent with expected random variation. A signal with a recurrence interval of 5000 days on the other hand has a small likelihood of occurring by chance alone (once every 14 years), and thus merits further investigation as an indication of a possible outbreak. High recurrence intervals may also be due to changes in hospital participation, specimen collection practices, or laboratory testing procedures. It is thus critical that statistical signals detected be further investigated through traditional epidemiological means.
WHONET-SaTScan analysis and signal consolidation
Analyses were performed using a specially adapted version of the WHONET 5.4 software which calls the SaTScan software from within. For each isolate analysed, the temporal data element was specimen collection date; the spatial data element was laboratory latitude and longitude, which were obtained using Google Earth [29]. Only patients' first isolates within a 60-day period were used in the analyses. In our simulated prospective system, we repeated the statistical analysis for each day of the surveillance period 1 July 2006 to 30 June 2007 using the previous 365 days as baseline. We used a maximum cluster length of 30 days and a maximum geographical size of 50% of the observations. The selection of parameters was supported by an optimization exercise (data not shown) which demonstrated that the events detected and conclusions drawn were robust with respect to a range of parameter settings. Recurrence intervals were calculated using 9999 Monte Carlo simulations.
In this study, we defined a signal worthy of further investigation as one with a recurrence interval of ⩾365 days thus ignoring signals consistent with chance occurrence more often than once a year. Consecutive daily signals were consolidated into ‘signal clusters’. There were many instances in which a signal cluster detected at one level of analysis, for example genus, overlapped in time and space with a signal cluster detected at another level of analysis, for example species or resistance phenotype. Such coincident clusters with overlapping isolates were grouped into epidemiological ‘events’ for purposes of interpretation. SaTScan-identified events and reported outbreaks were considered concordant if both implicated the same species of Shigella in the same province in the same month(s) and involved, when available, the same resistance phenotype and serotype.
Comparative data on shigellosis outbreaks
The national Ministry of Health (MoH) receives reports from provincial health departments on suspected and confirmed outbreaks of shigellosis and other foodborne diseases. Although outbreaks of shigellosis are not subject to mandatory public health reporting requirements in Argentina, local and provincial public health authorities believe that the extent of reporting of known outbreaks to the national level is relatively complete. We compared the statistically identified events from our system with the MoH outbreak registry by noting instances of overlap in time and space and drawing upon additional epidemiological and laboratory data (e.g. PFGE results, epidemiological notes) as available.
RESULTS
Event characterization
There were 2041 isolates analysed in the 12-month period. Fifty-three percent were in children aged <6 years, 20% in children aged 6–18 years, 8% in adults aged >18 years, and age was unknown in 19%. Frequencies of species and resistance phenotypes are presented in Table 1, while the seasonal variability of Shigella isolations is depicted in Figure 2. The most common species were S. flexneri (80%) and S. sonnei (17%).

Fig. 2. Frequency distribution of S. sonnei isolates non-susceptible to SXT by week for the laboratory in La Pampa associated with event 5. Isolates contributing to the SaTScan event are indicated by solid bars.
Table 1. Frequency of resistance phenotypesFootnote * of Shigella species isolated from specimens collected July 2006–June 2007
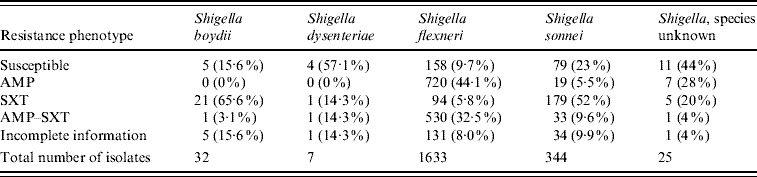
* Resistance profile definitions: Susceptible=Susceptible to AMP and SXT; AMP=non-susceptible to AMP, susceptible to SXT; SXT=susceptible to AMP, non-susceptible to SXT; AMP-SXT=non-susceptible to both AMP and SXT; Incomplete information=results for AMP and/or SXT are not available.
Our automated system found 31 statistical signal clusters grouped into 19 epidemiological events comprising a total of 187 isolates (Table 2, Fig. 3). The species most frequently associated with events were S. sonnei (47%) and S. flexneri (42%); there was one S. boydii event. The number of isolates in an event ranged from 2 to 55. Most events were associated with isolates reported by a single laboratory, while four events involved two or three laboratories – three events in Buenos Aires and one in two cities (Neuquén and Cipoletti) on opposite sites of the Limay River. An example of the temporal distribution of specimen collection dates showing both sporadic and clustered isolates is shown in Figure 4 for the laboratory involved in event 5.

Fig. 3. Location of outbreaks reported to the Ministry of Health (A–E) and statistical ‘events’ detected by SaTScan (1–19), July 2006–June 2007.

Fig. 4. Frequency distribution of Shigella spp. isolates included in the analyses by month July 2005 to November 2006 (n=29 laboratories); January 2006 to December 2007 (n=38 laboratories).
Table 2. Characteristics of statistical events detected by SaTScan, July 2006–June 2007
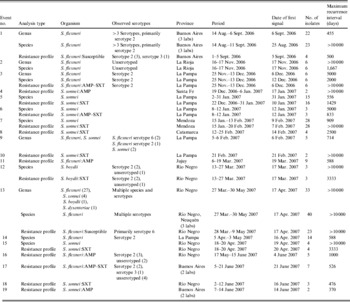
Given the clonal nature of most Shigella outbreaks, the most specific and epidemiologically meaningful analyses were those conducted with resistance phenotypes. Sixteen of the 19 events were identified in this way, and eight of these were seen only at this level. Two events were detected by species but not by resistance profile. In both of these, susceptibility testing was incomplete, precluding the possibility of detecting this outbreak by resistance profile.
Concordance between SaTScan and reported outbreaks
Outbreaks reported to the MoH during the July 2006–June 2007 surveillance period are shown in Tables 3 and 4 and Figure 3. Two of the six reported outbreaks were concordant, as defined in the Methods section, with SaTScan events: (1) outbreak F of S. flexneri serotype 6 in Río Negro in April with event 13; and (3) outbreak D of S. sonnei–SXT in La Pampa in January–February with events 5 and 10. Event 7 in the neighbouring province of Mendoza may have been related to outbreak D in light of a note recorded at the time by local laboratory staff indicating that the suspected source of the outbreak was produce originating in Mendoza.
Table 3. Shigellosis outbreaks reported to the Ministry of Health, July 2006–June 2007
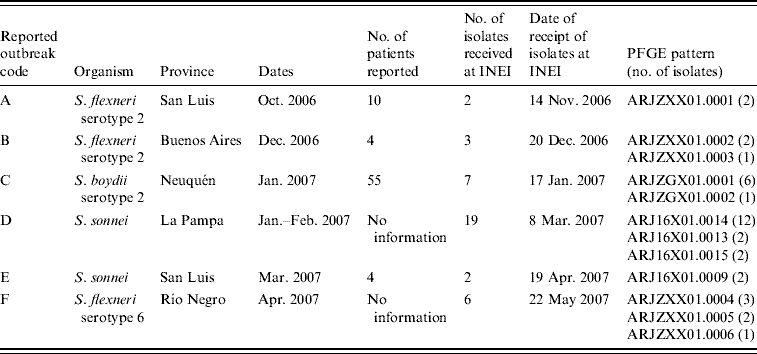
PFGE patterns were labelled according to guidelines of the PulseNet Latin America and Caribbean Network [Reference Swaminathan30]. The first two letters correspond to the country code (AR), then three characters for the bacterial species (JZX: S. flexneri; JZG: S. boydii; J16: S. sonnei), three for restriction enzyme (X01: XbaI) and finally the pattern number. For each outbreak listed, all isolates typed were considered to have highly similar PFGE patterns (up to two bands difference) and considered to be clonally related. PFGE patterns ARJ16X01.0014 (outbreak D) and ARJ16X01.0009 (outbreak E) were also highly related, with only two bands different.
Table 4. Shigellosis outbreaks reported to the Ministry of Health and concordant or possibly related events detected by SaTScan

Molecular typing demonstrated that outbreak E in March in San Luis (which did not contribute data for use in this study), which borders both Mendoza and La Pampa, was caused by the same strain as the one implicated in outbreak D. Thus, outbreaks D and E and events 5, 7, and 10 could reflect a single regional outbreak of S. sonnei non-susceptible to SXT involving La Pampa, Mendoza, and San Luis.
Outbreak C of S. boydii serotype 2, which took place in Neuquén in January among a group of students returning from a school trip to a distant city, was followed in March by event 12 just across the river in Cipolletti – three cases of the same species, serotype, and resistance pattern. S. boydii is relatively rare in Argentina (<2% of Shigella isolations) and had rarely been reported in Neuquén or Cipolletti previously, so the circumstances are suggestive, although certainly not conclusive, of some relationship between outbreak C and event 12 despite the several-weeks gap between the two. Local authorities felt that the two events were related although molecular confirmation was lacking.
In these simulated prospective analyses, SaTScan-generated signals appeared to be timely: local health authorities were notified of outbreak F on 19 April, 2 days after the first SaTScan-generated signal of event 13 in simulated prospective surveillance, and the first SaTScan signal of event 5 detected outbreak D on 10 January, a full 2 months prior to receipt on 8 March by INEI of the first isolates for confirmation.
Our analyses did not detect outbreaks A and B, and the relationship of three events with outbreak E was suggestive but not conclusive. Outbreaks A and E both occurred in San Luis, a centre excluded from the analyses presented in this study because of late enrolment in the network. The ability to detect outbreak B in Buenos Aires was probably compromised by the small number of patients (n=4) identified in a large urban area
Possible outbreaks identified by SaTScan not reported to the MoH
The epidemiological significance of the 14 SaTScan-identified events without evidence of a relationship to a known outbreak was further investigated through a review of additional details available in isolate line listings and discussions with local public health authorities. Isolate-level review suggests that 12 of these events were largely produced by clustering of a single Shigella strain, and thus were possibly associated with true outbreaks, while discordant isolate details (discrepant species, serotypes, and disk diffusion zone diameters) and low recurrence intervals suggest that events 9 and 19 were spurious statistical findings of low public health interest.
For 11 of these possible outbreaks, local authorities were not aware of any outbreaks with the characteristics indicated in Table 2. They pointed out that most of the events occurred during the summer months and that any excess isolates at the time were probably attributed to the usual expected rise in shigellosis cases typical of this time of year. Event 15 was interesting in that the reporting laboratory in Bariloche was very familiar with the set of patients identified – four patients with S. sonnei non-susceptible to SXT in a 3-day period which had struck local laboratory staff as unusual. A local investigation was initiated at that time. No epidemiological risk factors common to the patients were identified and local authorities concluded that this was not a public health outbreak and thus did not report this event to the MoH. This is an example of complete concordance between a SaTScan event and a known public health investigation, although the investigation concluded that this was not a true outbreak.
DISCUSSION
Public health strategies for the detection of outbreaks depend largely on non-statistical methods, ad hoc analyses, and unvalidated thresholds for action. Purely temporal statistics have been used effectively in many countries but with prior assumptions on geographic and temporal units studied and, in the case of pathogens with a strong seasonal component, requirements for several years of comparable baseline data. In this study, we have illustrated the potential use of electronic microbiology laboratory data for real-time disease outbreak detection, highlighting (1) the value of antimicrobial susceptibility test results in the detection of events of public health importance overlooked by current methods and (2) the benefit of space–time models, even when limited historical data are available. A focus on antimicrobial resistance phenotypes offers two important benefits. One is the possibility of supporting local and national resistance containment efforts through the early detection of and response to disease outbreaks due to antimicrobial-resistant pathogens and the prompt selection of appropriate antimicrobials during the outbreak period. The second potential benefit is to enhance the specificity and timeliness of outbreak detection by using the antimicrobial resistance phenotype as a marker to increase the statistical power for outbreak detection.
In this application of space-time outbreak detection algorithms for the detection of Shigella outbreaks utilizing the WHONET-Argentina dataset, we found good face validity in that at least two and possibly four of the six known Shigella outbreaks were found by our proposed system. Critically, our system detected an additional 14 events not reported to the MoH, most of which were suggestive of true outbreaks and certainly would have merited further epidemiological investigation and molecular characterization if detected in real time. Most of the suspected outbreaks reflected changes in the relative proportion and numbers of strains previously identified at a particular locale. However, some events (e.g. event 12: S. boydii in Río Negro; event 4: S. sonnei–AMP in Santa Fe) identified species or phenotypes rarely seen at those specific sites. The proposed system thus seems to have value not only in identifying outbreaks of frequently seen strains but also in the initial appearance of species and resistance phenotypes which are novel or infrequent at a particular location.
In this study, we used historical data to mimic a real-time prospective surveillance system. Such an approach can be used retrospectively as one measure of the effectiveness of public health surveillance and control programmes and also as part of an assessment of the burden of illness attributable to outbreaks. However, a real-time system with immediate confirmation and response would be of greater value,
The ability to implement such a real-time system depends on prompt data entry, analysis, and feedback as well as complete and consistent coverage of all provinces. A pilot project for prospective surveillance of Shigella outbreaks was launched in January 2009 for three contiguous provinces with weekly data submissions in close coordination with local authorities responsible for outbreak investigation and control. With the experience gathered in this pilot phase, prospective surveillance is likely to be extended to other pathogens and regions of the country.
The prospects for automating national laboratory-based surveillance for disease outbreak detection seem excellent. Our results highlight the value of antimicrobial resistance phenotypes in cluster detection both as a specific marker for outbreak detection and for early detection and containment of specific resistant pathogens of public health concern.
ACKNOWLEDGEMENTS
We extend our gratitude to all members of the Collaborative Group WHONET-Argentina and INEI staff for their ongoing efforts in support of the national surveillance programme. We thank Alejandra Corso, Allyson Abrams, Julie Dunn, and Gonzalo Vázquez-Prokopec for their important contributions to the research. We are also grateful to Susana Bruno and the Service of Antigens and Antisera of INPB-ANLIS ‘Carlos G. Malbrán’ for the provision of antisera for this study. Support was provided by cooperative agreement GM076672 from the National Institutes of Health under the Models of Infectious Disease Agent Study (MIDAS) programme and research project grant RR025040-02 from the National Institutes of Health.
APPENDIX
Collaborative Group WHONET-Argentina participants contributing data for this study : Buenos Aires (federal capital): P. O. Andrés*, D. Ballester*, E. N. Couto, A. L. Fernández*, N. A. Gómez, C. Lucero* N. Orellana, A. Procopio*, M. Quinteros, M. Vázquez*. Buenos Aires (province): R. Cabrera, B. Gatti*, D. Gómez, M. Machain*, A. Pacha, G. Páez*, S. Vaylet*, C. Vescina*. Catamarca: M. Ferres*, P. Váldez*, Córdoba: M. Bottiglieri, A. M. Littvik*, T. N. López*, L. Wolff De Jacob. Jujuy: S. Grosso, M. Toffoli, M. S. Weibel. La Pampa: G. Almada*, M. G. Gau De Cornejo*, N. Moreno, A. Pereyra. La Rioja: S. Flores De Galimberti*. Mendoza: L. Balbi*, M. A. Distefano, B. García*. Misiones: S. Grenón, A. M. Miranda, M. Von Specht. Neuquén: S. Brasili*, M. R. Núñez*, L. Pianciola*. Río Negro: N. Blázquez*, S. De Bunder*, M. C. Carranza*, N. Castro*. San Juan: H. Castro*, M. López, O. R. Navarro, R. Reinoso*. San Luis: E. M. Fernández*, Santa Cruz: H. Cano*, W. Krause*. Santa Fe: A. Badano*, N. Borda*, A. Ernst*, E. Méndez*, A. Mollerach*, R. Notario*. Santiago del Estero: M. Cragnolino, A. M. Nanni De Fuster. Tierra del Fuego: M. Vargas*, M. A. Laferrara*, Tucumán: M. A. Jure, H. Musa.
(* Denotes individuals from laboratories which have sent isolates to the Enteric Pathogens Service, INEI for serotyping and molecular studies.)
DECLARATION OF INTEREST
None.












