Implications
A substantial number of bulls whose semen passes the post-thaw quality control checks in artificial insemination (AI) centres have reduced fertility in the field. Although this is undoubtedly multifactorial, the purpose of this review is to attempt to identify where in the sequence of events, sperm from low-fertility bulls compromise the establishment of pregnancy. Understanding this will aid in the development of improved strategies for the early detection of bull subfertility and/or its amelioration.
Introduction
Animal breeding centres have for years relied upon classical microscopy-based techniques to assess sperm motility (total and progressive) and morphological parameters as part of their quality control programmes. However, work by our group and others have demonstrated that bulls whose semen passes these minimum post-thaw quality control checks at an AI centre can still vary in their field fertility. Traditional progeny testing schemes allowed semen from individual bulls to be released over a prolonged period and once non-return rate data became available, semen from sub-fertile bulls could be taken off the market. To protect against the risk of reduced fertility, AI companies typically utilise excessive sperm numbers in each straw (15 to 20 million). A number of studies with frozen-thawed conventional semen has revealed that most Holstein sires used in AI achieve their individual maximum pregnancy rate value at 2.5 to 5.0 million total sperm per dose, with a range from 0.5 to 12 million sperm per dose (Den Daas et al., Reference Den Daas, De Jong, Lansbergen and Van Wagtendonk-De Leeuw1998). Although the blanket approach of increasing the sperm number in all bulls guards against individual bulls with compensable sperm defects, this approach limits the number of straws that can be processed per ejaculate, thereby limiting supply of their semen.
With the advent of genomic selection, semen is now being collected from bulls at a younger age and these elite bulls are typically only used intensively for one season as they are then surpassed by the next generation of genetically superior bulls. This intensive use and high rate of AI sire turnover leaves insufficient time to adequately assess the fertility status of a bull before wide-scale use of his semen in the field, especially in seasonal grass-based production systems, such as those operated in Ireland and New Zealand. In these pasture-based systems, the breeding season is condensed into ~3 months so as to calve cows compactly at the start of the grass-growing season. These young bulls also produce fewer sperm per ejaculate, and therefore the luxury of putting excessive numbers of sperm in a semen straw for bulls where demand for semen far exceeds supply is costly for the AI centre and limits farmer access to such elite bulls. Therefore, a reliable in vitro test or a combination of tests, which could accurately predict the outcome of insemination would facilitate the identification of sub-fertile bulls before their widespread use in the field and the more efficient use of the semen of high fertility bulls through the reduction of sperm number per straw.
There are numerous recent studies and comprehensive reviews on the prediction of bull fertility (Sellem et al., Reference Sellem, Broekhuijse, Chevrier, Camugli, Schmitt, Schibler and Koenen2015; Utt, Reference Utt2016; Abdollahi-Arpanahi et al., Reference Abdollahi-Arpanahi, Morota and Penagaricano2017) and we do not propose to replicate these here. Instead, this review will focus on the caveats surrounding sire fertility estimates and on the specific reasons why bulls with apparently normal semen vary in their fertility. We assess the usefulness of in vitro assessments to mimic the in vivo events leading up to the establishment of a pregnancy. Finally, we propose likely avenues for fruitful future investigation.
Sire fertility estimates; establishing a reliable phenotype
Many studies that use in vitro approaches to investigate bull fertility fail to understand the limitations of even the best designed sire fertility estimates and thus many studies are flawed from the start due to an unreliable fertility phenotype. To accurately rank sires, a detailed understanding of factors affecting the models are required. Most AI centres worldwide track bull fertility using either non-return rates or more accurate (and complex) adjusted sire conception rate (SCR) models that account for environmental factors (herd, technician, month of insemination, age of cow, cow genotype, days in milk, milk production, etc.) and express a bull’s fertility relative to a population mean of 0%. A detailed review by Amann and DeJarnette (Reference Amann and DeJarnette2012) demonstrated that the fertility of 90% of the bulls marketed is within±3 percentage points of the mean of the bull population. This is consistent with our preliminary data on Irish AI bulls. On a population basis, Amann and DeJarnette (Reference Amann and DeJarnette2012) concluded that AI companies will never be able to measure ‘fertility’ more precisely than ±3 percentage units from the population mean because of the difficulty in controlling many factors including: binomial variation, herd environment, measurement errors, and bias in semen use. Another key attribute in understanding ‘Sire Fertility’ is the number of inseminations required per sire to confidently rank them on their fertility. The same study illustrated that to confidently (two tailed test, P=0.05, 80% power) differentiate sires ±4% from the average of the bull population each sire must have a minimum of 1000 inseminations. With just 300 inseminations, as is often the case in studies attempting to predict the fertility of an individual ejaculate, it is only possible to confidently differentiate sires±7% from the average. Thus, failure to recognise limitations in any estimate of potential fertility leads to over interpretation of small differences among sires in apparent fertility.
Do artificial insemination bulls rank the same when used under different conditions?
Although the timing of insemination relative to onset of oestrus does not influence the fertility of above average fertility sires, a significant drop in fertility was reported when semen from below average sires was inseminated in early and mid-oestrus (Macmillan and Curnow, Reference Macmillan and Curnow1977), suggesting differences in the fertile lifespan of sperm in the female tract. Despite this fact, once a day AI is now widely used with similar fertility achieved to when twice a day AI is performed, irrespective of whether fresh or frozen-thawed semen is used (Xu, Reference Xu2017).
Optimum fertility can be achieved with a much lower sperm number when fresh (liquid) rather than frozen-thawed semen is used (2 to 5 million v. 15 to 20 million sperm, respectively; Murphy et al., Reference Murphy, Eivers, O’Meara, Lonergan and Fair2017), maximising the utilisation of genetically superior sires. The higher sperm numbers in cryopreserved semen compensate for the damage during the freeze-thaw process compared with fresh semen and, on average, the same level of fertility is achieved with both types of semen (Murphy et al., Reference Murphy, Holden, Murphy, Cromie, Lonergan and Fair2015). In a data set analysed by our group, bulls which have low fertility with frozen-thawed semen tend to have low fertility with fresh semen although there are some exceptions (Figure 1). Of the 16 bulls used across 66 252 inseminations, nine bulls varied substantially in the fertility achieved between fresh and frozen-thawed semen (five bulls were higher with fresh semen and four were higher with frozen-thawed). In contrast, Vishwanath and Shannon (Reference Vishwanath and Shannon2000) reported that bulls generally followed the same fertility trend with fresh and frozen-thawed semen when optimum sperm numbers were used (2.5 million/dose for fresh and 20 million/dose for frozen-thawed) but when suboptimum sperm numbers were used (0.5 million/dose for fresh and 5 million/dose for frozen-thawed) fertility declined by 7% and 7.9% for fresh and frozen-thawed, respectively. More importantly, there was a significant bull by sperm number interaction, whereby some bulls dropped by over 20% when lower sperm numbers of frozen-thawed semen was inseminated and some bulls performed the same as with 20 million sperm. This trend of a greater variation among bulls at lower sperm concentrations was also observed by Den Daas et al. (Reference Den Daas, De Jong, Lansbergen and Van Wagtendonk-De Leeuw1998) where maximum fertility for individual bulls was achieved at differing sperm concentrations, and the sperm numbers needed to obtain 95% of the maximal conception rate ranged from 1 to 11 million sperm per dose. This highlights the variability in the susceptibility of an individual bull’s semen to the freeze-thaw process and how freezing protocols should be customised to individual bulls, an area of research that has received little attention in recent years. It also illustrates that individual bulls have different maximum fertility and the number of sperm required to achieve this varies among bulls.
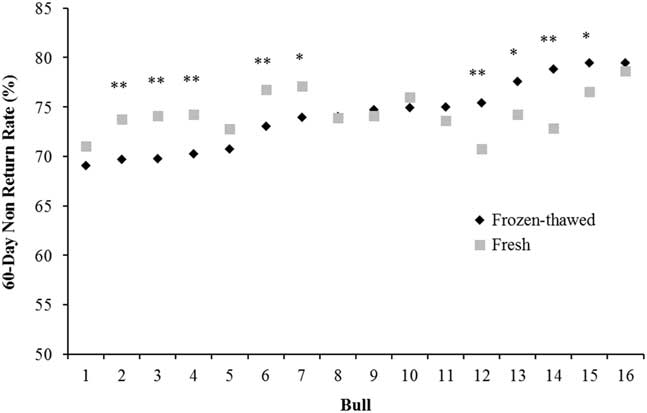
Figure 1 Characterisation of the variation between 60 day non-return rate in 16 bulls with split ejaculates used as fresh or frozen-thawed semen (n=66 252 inseminations). *P<0.05, **P<0.01 between semen type within bull.
In order to minimise differences in pregnancy rates among individual sires, over-compensation of sperm numbers typically occurs in the preparation of frozen-thawed semen, resulting in a sperm concentration that considerably exceeds the number of sperm necessary for maximum fertility. Thus, the ‘true fertility’ potential of a bull in the field is masked by the greater sperm number per insemination dose, and this needs to be considered when attempting to understand the variation in SCR using in vitro assays. For example, consider an AI centre which processes semen at 15 million sperm per inseminate (as is typical) with an overall mean calving rate across all its bulls of 53%. A comparison of two of their bulls with a calving rate of 60% would lead to the conclusion that both bulls were of ‘high fertility’ and in any retrospective ‘prediction type analysis’, they would be treated as such. However, now consider that if assessed at a lower sperm number, one bull would have had the same fertility at a dose of 5 million sperm per straw while the second bull would have required 12 million sperm for this level of fertility. As the only fertility data available to the AI centre was at a concentration of 15 million sperm, both bulls would be considered to have the same fertility phenotype yet there are distinctive differences in the ability of their sperm to establish a pregnancy after the freeze-thaw process. Therefore, fertility can only be actually determined under conditions where sperm numbers are limiting (Hammerstedt, Reference Hammerstedt1996) and it is not surprising that there is difficulty in identifying the causes of bull subfertility when a dubious fertility phenotype is used at the start. Ideally, the number of sperm at which a bull’s fertility reaches a plateau should be determined, but tracking semen straws with varying sperm concentrations in the field poses major logistical issues for most AI companies.
Where does reproductive wastage occur in bulls that vary in their sire conception rate following artificial insemination?
There have been a plethora of publications on the prediction of sire fertility using sperm functional (Sellem et al., Reference Sellem, Broekhuijse, Chevrier, Camugli, Schmitt, Schibler and Koenen2015), molecular (Rahman et al., Reference Rahman, Kwon and Pang2017) and genomic (Puglisi et al., Reference Puglisi, Gaspa, Balduzzi, Severgnini, Vanni, Macciotta and Galli2016) models as well as combinations of these. Despite this, there is still no single test, or combination of tests, which can reliably predict bull fertility. Very few studies have focused on attempting to understand why bulls whose semen has normal post-thaw motility and morphology, as viewed under a microscope, can still vary in fertility by up to 20% points. For most commercial situations, fertility is defined as cows either failing to return to oestrus (non-return rate) or confirmed pregnant by means of ultrasound scan, rectal palpation, blood progesterone or a calving event. These estimates of pregnancy status following insemination are incapable of differentiating the reasons for pregnancy failure. What is clear is that semen is deposited into the uterine body and the chance of pregnancy varies among bulls. The possible reasons for this are presented in Figure 2 and the associated published studies are then discussed.
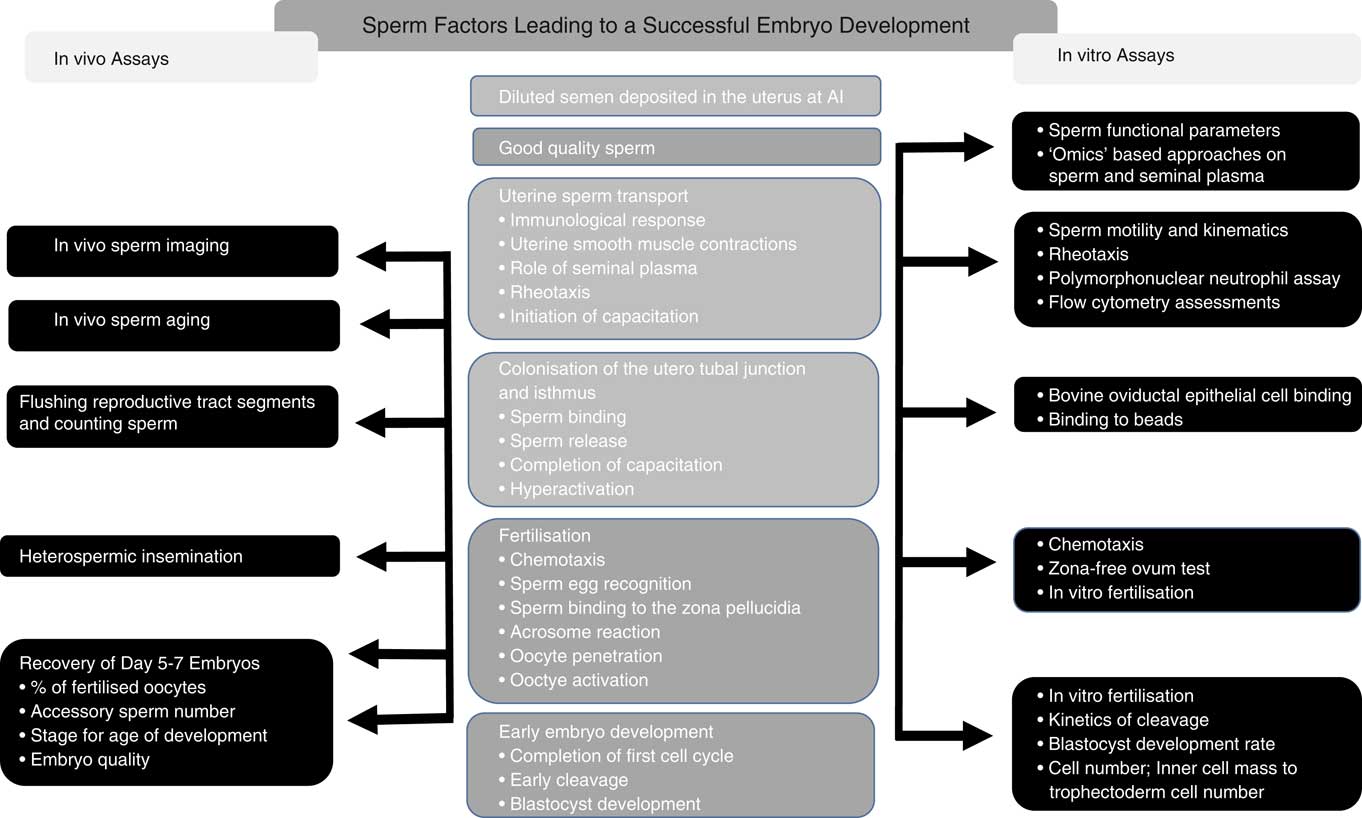
Figure 2 Events leading to the establishment of a viable embryo following artificial insemination (AI; centre column), with the in vivo (left column) as well as the in vitro (right column) assessments that have been used in published studies to characterise the differences in these events between bulls of varying fertility.
Sperm proteome and its relationship to the establishment of pregnancy
During ejaculation, sperm becomes coated in proteins immediately during ejaculation that is secreted from the epididymides as well as the accessory glands and even though bull semen is typically diluted 15 to 25 fold during semen processing, the effects of seminal plasma proteins are likely to be maintained as they adhere to sperm rapidly upon ejaculation. Numerous studies have focused on characterising the proteomic composition of the seminal fluid (which also contains epididymal fluid) across a range of species (Druart et al., Reference Druart, Rickard, Mactier, Kohnke, Kershaw-Young, Bathgate, Gibb, Crossett, Tsikis, Labas, Harichaux, Grupen and de Graaf2013) and related these to fertility. Some of the seminal plasma proteins that have been positively related to bull fertility include osteopontin (Ca2+-binding protein) and lipocalin-type prostaglandin D synthase (Cancel et al., Reference Cancel, Chapman and Killian1997), telomeres-1 protein (POT1) (Aslam et al., Reference Aslam, Kumaresan, Sharma, Tajmul, Chhillar, Chakravarty, Manimaran, Mohanty, Srinivasan and Yadav2014) while other seminal plasma proteins have been negatively correlated to fertility including prostaglandin E2 receptor EP3 (PTGER3) (Aslam et al., Reference Aslam, Kumaresan, Sharma, Tajmul, Chhillar, Chakravarty, Manimaran, Mohanty, Srinivasan and Yadav2014). More functional studies are required to validate these and to characterise how exactly they influence the establishment of pregnancy.
Other studies have mapped the proteome of bull sperm and have reported correlations between specific proteins and sperm motility, morphology as well as fertility (D’Amours et al., Reference D’Amours, Frenette, Fortier, Leclerc and Sullivan2010). Sperm proteins can be broadly categorised into energy-related, structural and other functional proteins and sperm-bound proteins from bulls of varying fertility have been related to spermiation and energy homoeostasis, membrane function, sperm-egg interactions and cell cycle regulation as well as glycolysis, post-translational changes during sperm maturation, capacitation and protection against oxidative stress, to name but a few (Gaviraghi et al., Reference Gaviraghi, Deriu, Soggiu, Galli, Bonacina, Bonizzi and Roncada2010; Park et al., Reference Park, Kwon, Oh and Pang2012). Molecular defects in some of these proteins have been reported to be associated with low fertility or in certain cases, infertility. Somashekar et al. (Reference Somashekar, Selvaraju, Parthipan and Ravindra2017) investigating sperm proteomic signatures regulating sperm function and fertility reported calmodulin (CALM1), spermadhesinZ13 (SPADH2), and phosphatidylethanolamine-binding protein 4 (PEBP4) to be present in higher amounts on the sperm of high fertility bulls with PEBP4 being absent in infertile bulls. An earlier study by the same group reported that the seminal plasma protein PDC-109 was more abundant on sperm from low-fertility bulls (Somashekar et al., Reference Somashekar, Selvaraju, Parthipan, Patil, Binsila, Venkataswamy, Karthik Bhat and Ravindra2015). The exact role of many of these proteins in bull fertility is still unclear, and thus the current challenge for reproductive biologists is to move from lists of identified proteins to an informed understanding of biological function given that they control key physiological events in the female tract.
Sperm communication and interaction with the female tract
The immunological responses to sperm and seminal plasma in the female tract are of considerable interest as these processes influence sperm capacitation, transport, selection, fertilisation as well as early embryo development (Schuberth et al., Reference Schuberth, Taylor, Zerbe, Waberski, Hunter and Rath2008). The local immune responses of the epithelial lining, regulated by its secretions, constitute the main part of the mucosal innate immunity inside the uterus and oviduct which is largely mediated by cytokines, chemokines, and prostaglandins (Bulek et al., Reference Bulek, Swaidani, Aronica and Li2010). Much of the focus in humans and rodents have been on the bioactive signalling agents in seminal plasma and how they evoke gene expression and cellular changes in the innate immune system (see review by Schjenken and Robertson, Reference Schjenken and Robertson2014). The presence of sperm, seminal plasma and semen diluent causes a triggering of the first line of defense against foreign cells through increased production of pro-inflammatory cytokines, leading to an influx of polymorphonuclear neutrophils (PMNs) into the lumen of the female tract (Marey et al., Reference Marey, Yousef, Liu, Morita, Sasaki, Hayakawa, Shimizu, Elshahawy and Miyamoto2014). Polymorphonuclear neutrophils have been reported to clear dead and immotile sperm, but also motile sperm (Li and Funahashi, Reference Li and Funahashi2010) and the presence of activated phagocytes can lead to decreases in sperm motility due to increased production of reactive oxygen species (ROS). Oxidative stress is known to be a major factor regulating the vitality and functionality of sperm; however, the precise implications of increased ROS within the female tract are not well understood. Sperm from bulls with below average fertility had significantly greater ROS production compared with above average fertility bulls (Kumaresan et al., Reference Kumaresan, Johannisson, Al-Essawe and Morrell2017) which has in turn been related to increased deleterious effects of lipid peroxidation on the membrane and DNA integrity (Koppers et al., Reference Koppers, Mitchell, Wang, Lin and Aitken2011).
Using an in vitro model, Marey et al. (Reference Marey, Liu, Kowsar, Haneda, Matsui, Sasaki, Takashi, Hayakawa, Wijayagunawardane, Hussein and Miyamoto2016) demonstrated that endothelin-1 may be involved in supporting bull sperm survival until fertilisation through the protection of sperm from phagocytosis by PMNs in the bovine oviduct. In the macaque, beta-defensin 126 (BD126) has been reported to protect sperm from immune-recognition and binding of anti-sperm antibodies (ASA; Yudin et al., Reference Yudin, Generao, Tollner, Treece, Overstreet and Cherr2005). Anti-sperm antibodies are produced in response to antigens present on sperm and can account for reduced sperm viability but also higher sperm mortality in the female reproductive tract (Rossato et al., Reference Rossato, Galeazzi, Ferigo and Foresta2004). It is estimated that ASA are responsible for as much as 40% of unexplained fertility cases in humans and recent studies have also revealed a high level of ASA in both serum and seminal plasma from bulls that have a negative effect on their fertility through the prevention of capacitation (Zodinsanga et al., Reference Zodinsanga, Cheema and Mavi2015).
β-defensin glycoproteins coat sperm and have been identified as having a role in modulating the inflammatory response to enhance sperm survival (Yudin et al., Reference Yudin, Generao, Tollner, Treece, Overstreet and Cherr2005). Although the function of β-defensins in reproduction have not, until recently, been explored in farm animal species, knockout of a β-defensin gene cluster in male mice resulted in complete sterility (Zhou et al., Reference Zhou, Webb, Lettice, Tardif, Kilanowski, Tyrrell, Macpherson, Semple, Tennant, Baker, Hart, Devenney, Perry, Davey, Barran, Barratt and Dorin2013). In men, variation in the bd126 sequence contributes to subfertility (Tollner et al., Reference Tollner, Yudin, Tarantal, Treece, Overstreet and Cherr2011) while the BD126 peptide has been reported to mediate sperm binding to the oviductal epithelium (Tollner et al., Reference Tollner, Venners, Hollox, Yudin, Liu, Tang, Xing, Kays, Lau, Overstreet, Xu, Bevins and Cherr2008). Our group has reported bovine beta-defensin 126 (BBD126) to be extensively expressed in the reproductive tract of the bull with preferential protein expression in the cauda epididymis (Narciandi et al., Reference Narciandi, Fernandez-Fuertes, Khairulzaman, Jahns, King, Finlay, Mok, Fair, Lonergan, Farrelly and Meade2011) and on sperm (Narciandi et al., Reference Narciandi, Lloyd, Chapwanya and Meade2016) with similar binding patterns on the sperm surface to macaque. Beta-defensin 126 increases the net negative charge on sperm (Tollner et al., Reference Tollner, Bevins and Cherr2012), increases sperm motility, mucus penetration in vitro (Fernandez-Fuertes et al., Reference Fernandez-Fuertes, Narciandi, O’Farrelly, Kelly, Fair, Meade and Lonergan2016) as well as sperm binding to oviductal epithelium in vitro (Lyons et al., Reference Lyons, Narciandi, Donnellan, Romero-Aguirregomezcorta, O’Farrelly, Lonergan, Meade and Fair2018). We have also recently characterised the genetic variation in bovine β-defensin genes as well as completing the first whole-exome sequencing of AI bulls of divergent fertility (Whiston et al., Reference Whiston, Finlay, McCabe, Cormican, Flynn, Cromie, Hansen, Lyons, Fair, Lonergan, O’ Farrelly and Meade2017). This dual approach successfully identified novel variants in both beta-defensin and FOXJ3 genes as potentially regulating SCR through differential oviductal binding ability, as assessed in vitro (Whiston et al., Reference Whiston, Finlay, McCabe, Cormican, Flynn, Cromie, Hansen, Lyons, Fair, Lonergan, O’ Farrelly and Meade2017). Using a microarray-based approach, Legare et al. (Reference Legare, Akintayo, Blondin, Calvo and Sullivan2017) characterised the expression of genes along the caput, corpus and cauda epididymis in bulls which differed in SCR. The transcriptional profiles between sub-fertile and fertile bulls clustered most closely in the cauda and corpus segments, whereas the profiles in the caput segment were distinct between sub-fertile and fertile bulls. Of the differently expressed genes, 10 were related to reproductive function and five were associated with the defense response (of which two belonged to the defensin family, namely DEFB119, DEFB124). Bulls carrying mutations in genes which encode these immunoregulatory peptides could produce sperm of higher immunogenicity which could well contribute to reduced sperm survival in the female reproductive tract and subfertility.
During ejaculation, a binder of sperm proteins (BSPs) are secreted by bovine seminal vesicles into seminal plasma and immediately absorbed onto sperm (Leahy and de Graaf, Reference Leahy and de Graaf2012). Of these, BSP1, BSP3, BSP5 have been reported to facilitate uncapactitated bull sperm in binding to the epithelial lining of the utero-tubular junction and isthmus, forming a sperm storage reservoir (Hung and Suarez, Reference Hung and Suarez2012). However, during the completion of capacitation, changes in their composition on sperm play a role in releasing sperm from these storage reservoirs. The ability of sperm to bind to the oviductal epithelium appears critical to establishing a viable sperm population in the oviducts and may aid in overcoming any asynchrony between the timing of AI and ovulation. A number of studies have investigated the interaction between sperm and the oviductal epithelium in vitro and demonstrated that it is mediated by fucose (Lefebvre et al., Reference Lefebvre, Lo and Suarez1997). Previous in vivo work has reported that the timing of insemination is more important for low and average fertility bulls compared with high fertility bulls (Macmillan and Watson, Reference Macmillan and Watson1975) suggesting that there is a reduced ability of sperm from low-fertility bulls to develop a reservoir of functional sperm at the utero-tubular junction and in the oviducts. Interestingly, Yousef et al. (Reference Yousef, Marey, Hambruch, Hayakawa, Shimizu, Hussien, Abdel-Razek, Pfarrer and Miyamoto2016) reported that bovine oviductal epithelial cells provide an anti-inflammatory environment and the sperm-epithelial binding further strengthens this, leading to the suppression of PMNs in the bovine oviduct. In addition to facilitating sperm binding, Lessard et al. (Reference Lessard, Siqueira, D’Amours, Sullivan, Leclerc and Palmer2011) investigating the aetiology of idiopathic infertility in a beef bull established that his sperm were unable to undergo the acrosome reaction, when induced using calcium ionophore, and related this to the level of BSP1 that was much greater on sperm from the infertile bull compared with that of his sire.
It is clear that there is cross-talk between semen and the female tract, starting in the uterus with the induction of an inflammatory response and continuing in the oviduct through sperm binding and subsequent release. There is evidence that bulls vary in their capacity to complete these physiological processes and indications are that this is related to the surface proteome of sperm that is influenced by both the epididymal secretome and the composition of seminal plasma. The focus, therefore, should be on characterising these parameters from bulls of divergent fertility with a view to identifying key biomarkers (not just proteins), which can then be used in functional studies to better understand how these regulate the dialogue between sperm and female reproductive tract.
Sperm transport in the female tract
In vivo assessments, either by flushing sperm from the segments of the reproductive tract following AI or using confocal imaging of sperm in the female tract as has been performed in sheep (Druart et al., Reference Druart, Cognie, Baril, Clement, Dacheux and Gatti2009), are unlikely to be sensitive enough to detect differences among bulls with varying SCR. Other approaches such as mucus penetration tests, assessment of accessory sperm number following AI and heterospermic insemination have been used to understand why some males sperm may be better able to navigate the female reproductive tract and its secretions than others.
The use of mucus penetration assays in vitro, which assess the ability of the sperm to travel through a capillary filled with artificial mucus or cervical mucus from oestrus cows has been used as a proxy for assessment of sperm transport and has been correlated to SCR (Al Naib et al., Reference Al Naib, Hanrahan, Lonergan and Fair2011). Although these studies were conducted in a static mucus environment, the development of microfluidic systems enables the characterisation of sperm rheotaxis, a phenomenon whereby sperm swim against a flow (Miki and Clapham, Reference Miki and Clapham2013) and offers a way of assessing sperm migration ability. Although chemotaxis may guide sperm towards the ovulated oocyte once it is in its vicinity in the ampulla, rheotaxis has been proposed as a long-range guidance cue for sperm navigation along the female tract. Rheotaxis requires rotation of the sperm, which requires CatSper calcium-selective ion channels. CatSper glycoproteins form the sperm-specific voltage-gated Ca2+ channels localised along the membrane of the sperm flagellum. CatSper channels contain glycoproteins that are involved in positioning regulation and recent work by our group has demonstrated that hyperactive bull sperm exhibit an increased rheotaxis response (Johnson et al., Reference Johnson, English, Cronin, Hoey, Meade and Fair2017). Targeted disruption of CATSPER 1, CATSPER2, CATSPER3 or CATSPER4 inhibits hyperactivated motility and thus rheotactic reponse (Johnson et al., Reference Johnson, English, Cronin, Hoey, Meade and Fair2017). There are no published studies on the rheotactic response of bulls differing in SCR. The advent of 3-D printing will no doubt facilitate the development of more physiological and sensitive models for studying this as well as sperm interaction with the female tract and its secretions.
Heterospermic insemination involves the insemination of the semen mixture at the one point in time into the same female and thus levels the playing field. Each sperm should have an equal chance to reach and fertilise the oocyte without influence by technician, cow age/parity/genetic merit/days in milk, timing of insemination relative to oestrus, season and management (Beatty et al., Reference Beatty, Bennett, Hall, Hancock and Stewart1969). For this reason, it has been reported that heterospermic insemination is up to 170 times more sensitive in ranking reproductive outcome than homospermic measures (Flint et al., Reference Flint, Chapman and Seidel2003). To put this into context, homospermic insemination requires thousands of inseminations to compare fertility of two males accurately while heterospermic insemination has been reported to be able to test the fertility of a bull accurately and rapidly using fewer than 100 females. Overstreet and Adams (Reference Overstreet and Adams1971) inseminated a mixture of equal numbers of labelled and unlabelled rabbit sperm from two bucks and flushed the reproductive tract of does 6 or 13 h later for evidence of selective transport and sperm viability. The numbers of sperm from each male in each of the segments of the reproductive tract were equal at 6 h, but by 13 h sperm from the superior buck predominated in the uterus and oviducts. More sperm from the superior buck were attached to the zona pellucida and fertilised more oocytes. When semen was placed in the oviducts the sperm were present in equal numbers in the vicinity of the oocyte but the skewed proportion of offspring and labelled sperm penetrating the oocyte still favoured the superior buck. Using heterospermic insemination of fluorescently labelled sperm, Ferreira (Reference Ferreira1972) reported that sperm number recovered from the vagina, uterus and oviduct was similar among males, as was the number of sperm bound to the zona pellucida of recovered oocytes. These observations lead the authors of these aforementioned studies to conclude that sperm are present in equal numbers in the immediate vicinity of the oocyte and perhaps rate of oocyte penetration or subsequent activation of the oocyte differed among males. This is also in agreement with Macmillan and Watson (Reference Macmillan and Watson1975) who reported that all bulls have a similar opportunity to fertilise when AI occurred close to ovulation, but when AI occurred at longer intervals before ovulation, the sperm of some bulls, which were obviously present at longer intervals to AI, were no longer alive or capable of fertilising an oocyte. These studies emphasise the importance of having a population of functional sperm in the oviducts at the time of ovulation.
Fertilisation and early embryo development
Fertilisation success following AI in cattle with semen from high fertility sires is in the order of 90% to 95% in heifers and moderate yielding cows (Diskin and Sreenan, Reference Diskin and Sreenan1980). A meta-analysis by Sartori et al. (Reference Sartori, Bastos and Wiltbank2010) estimated that fertilisation rates in North American high-producing Holstein cows to be 83% while pregnancy rates of similar genetic merit lactating cows to be 33%. Several studies have been performed over the last 30 years in which cows have been slaughtered at various time-points post insemination in order to assess embryo viability. The majority of this reproductive wastage in single-ovulating cows had been attributed to early embryo loss with <50% of recovered embryos from high yielding lactating cows viable 7 days after AI followed by additional losses through Day ~34 (Sreenan and Diskin, Reference Sreenan and Diskin1986). Sartori et al. (Reference Sartori, Sartor-Bergfelt, Mertens, Guenther, Parrish and Wiltbank2002) demonstrated that lactating cows had poorer quality Day 5 embryos than both heifers and dry cows but surprisingly more accessory sperm indicating that delayed sperm transport was not a causative effect. In single-ovulating cows, most embryos and ~80% of unfertilised oocytes had at least one accessory spermatozoon (Cerri et al., Reference Cerri, Rutigliano, Lima, Araujo and Santos2009) while Sartori et al. (Reference Sartori, Sartor-Bergfelt, Mertens, Guenther, Parrish and Wiltbank2002) reported mean values of 18–42 sperm in embryos and 18 in unfertilised ova. All of these aforementioned studies were focused on cow factors and there is a complete dearth of published studies focusing on the relationship between SCR and the contribution of the sperm to failure of sperm transport, fertilisation or embryo development. Using a small number of bulls with below and above average fertility, Ortega et al. (Reference Ortega, Moraes, Patterson, Smith, Poock and Spencer2017) recently assessed the contribution SCR to pregnancy establishment and reported that bulls with a higher SCR had an advantage in terms of in vivo and in vitro production of embryos. In the same study there was no effect of SCR on preimplantation conceptus elongation and development. Kumaresan et al. (Reference Kumaresan, Johannisson, Al-Essawe and Morrell2017) reported that bulls with below average fertility had a significantly lower sperm population with intact acrosomes post-thawing compared with bulls with above average fertility, similar to earlier reports (Singh et al., Reference Singh, Kumaresan, Chhillar, Rajak, Tripathi, Nayak, Datta, Mohanty and Malhotra2016).
In vitro fertilisation (IVF) is a powerful tool to assess the fertilising ability of sperm. The kinetics of sperm penetration (Ward et al., Reference Ward, Enright, Rizos, Boland and Lonergan2002) as well as the first cell cycle (Comizzoli et al., Reference Comizzoli, Marquant-Le Guienne, Heyman and Renard2000) and of the first mitotic cleavage after fertilisation (Lonergan et al., 1999) are highly correlated with the likelihood of an embryo developing to the blastocyst stage and to the quality of those embryos (Dinnyes et al., Reference Dinnyes, Lonergan, Fair, Boland and Yang1999). Ward et al. (Reference Ward, Rizos, Corridan, Quinn, Boland and Lonergan2001) was able to discriminate between bulls of high and low field fertility based on the timing of the first cleavage division post insemination in vitro, whereby embryos fertilised from high fertility bulls cleaved first and significantly more of these early cleaving zygotes were more competent in terms of development to the blastocyst stage than those that cleaved later. The same study reported a significant correlation between Day 7 blastocyst yield and field fertility while a separate study reported an effect of SCR and cleavage rate (Al Naib et al., Reference Al Naib, Hanrahan, Lonergan and Fair2011). In contrast, Kropp et al. (Reference Kropp, Carrillo, Namous, Daniels, Salih, Song and Khatib2017) reported no differences in the morphology and development to the blastocyst stage but preimplantation embryos derived from high and low-fertility bulls displayed significant transcriptomic differences, which they postulated could influence the reprogramming of the early embryo. Therefore, the evidence suggests that a portion (contribution will vary among sires) of the embryo death before ~Day 8 is caused by the fertilising sperm, but the specific aspect of the sperm causing this effect is unclear.
Role of sperm DNA integrity and methylation signature
Individual bulls vary in the levels of sperm DNA fragmentation that they exhibit (Takeda et al., Reference Takeda, Uchiyama, Kinukawa, Tagami, Kaneda and Watanabe2015) and there appears to be a growing link between this parameter and early embryonic loss and even foetal development and health of the offspring (Evenson and Jost, Reference Evenson and Jost2000). During spermiogenesis, sperm chromatin is remodelled whereby core histones are replaced by transition proteins which are subsequently replaced by protamines resulting in chromatin that is tightly compacted and resistant to denaturation (Filho et al., Reference Filho, Beletti and de Oliveira2015). This compaction is necessary to protect sperm chromatin during transit through the epididymis and female reproductive tract. Shortly after fertilisation, sperm protamines are replaced by maternal histone variants. Thus, defects of sperm chromatin structure affect sperm function during fertilisation, first cleavage and early embryonic development. Inadequate sperm chromatin protamination and DNA integrity were associated with defects in bull sperm chromatin condensation, coinciding with reduced in vivo fertility (Dogan et al., Reference Dogan, Vargovic, Oliveira, Belser, Kaya, Moura, Sutovsky, Parrish, Topper and Memili2015). Disruption to defective chromatin packaging during spermiogenesis results in sperm that are susceptible to denaturation and there is growing evidence, that the status of sperm chromatin at the time of fertilisation can influence embryonic survival (Sakkas et al., Reference Sakkas, Moffatt, Manicardi, Mariethoz, Tarozzi and Bizzaro2002).
A number of studies have demonstrated a relationship between the levels of DNA fragmentation in bull sperm and SCR (Kumaresan et al., Reference Kumaresan, Johannisson, Al-Essawe and Morrell2017) with a high level of DNA fragmentation correlated to sperm morphology (Nagy et al., Reference Nagy, Johannisson, Wahlsten, Ijas, Andersson and Rodriguez-Martinez2013) as well as to a reduced sperm fertilisation potential. DNA fragmentation values of between 7% and 10% have been reported to be indicative of low AI success in bulls as DNA damage can jeopardise embryonic development (Karoui et al., Reference Karoui, Díaz, González-Marín, Amenabar, Serrano, Ugarte, Gosálvez, Roy, López-Fernández and Carabaño2012). However, it must also be noted that many bulls that have lower fertility do not always exhibit increased levels of DNA fragmentation as Rodriguez-Martinez and Barth (Reference Rodriguez-Martinez and Barth2007) reported no direct correlation of DNA fragmentation with fertility. Therefore, like many other in vitro parameters, DNA fragmentation seems more useful when using the negative biomarker approach whereby high levels indicate sperm defects, but low levels do not guarantee fertility.
It is has been known for some time that, at the time of fertilisation, sperm deliver much more than just DNA, but rather an entire package including RNAs, transcription factors, and cell signalling molecules (Krawetz, Reference Krawetz2005). Although a number of studies have demonstrated that the transcriptome is significantly different among sires of varying fertility (Feugang et al., Reference Feugang, Rodriguez-Osorio, Kaya, Wang, Page, Ostermeier, Topper and Memili2010), it has only recently been reported that the embryonic transcriptome is influenced by the ‘RNA package’ delivered by sires of varying fertility status at the time of fertilisation (Kropp et al., Reference Kropp, Carrillo, Namous, Daniels, Salih, Song and Khatib2017). The same study characterised the epigenetic signature of the sperm between bulls of high and low fertility and revealed 76 regions to be differentially methylated between sires of divergent fertility. Although cleavage and blastocyst rate was not affected, the resultant IVF-derived embryos had significantly different transcriptomic profiles with genes relating to metabolic processes and catalytic activities more highly expressed in sperm from high fertility bulls. Errors relating to the condensation of the DNA during spermatogenesis as well as maintenance of epigenetic marks could possibly explain the differences in embryonic gene expression. Indeed, lower levels of DNA condensation, protamine exchange, and higher DNA damage have been observed in sperm from lower fertility bulls in comparison to higher fertility bulls (Dogan et al., Reference Dogan, Vargovic, Oliveira, Belser, Kaya, Moura, Sutovsky, Parrish, Topper and Memili2015). In addition, a recent study focusing on the epigenetic profiles of young bulls highlighted that 10-month-old bulls have a different sperm DNA methylation pattern compared with both 12- and 16-month-old bulls (Lambert et al., Reference Lambert, Blondin, Vigneault, Labrecque, Dufort and Sirard2018). Given the current trend of using semen from elite genomically selected bulls, this study demonstrates that such bulls not only have poorer sperm motility and morphology but also an altered epigenetic profile that has the potential to influence embryonic development as well as the genotype and the phenotype of the subsequent offspring.
Sperm RNA and its relationship to sire conception rate
Ejaculated sperm are ‘stripped-down’ cells, equipped with a strong flagellum to drive them through an aqueous mucus environment but unencumbered by cytoplasmic organelles. As a result they are transcriptionally inactive but do retain remnant messenger RNA (mRNA) that are left over from spermatogenesis that can be used for diagnostic purposes. Thus, transcripts (and translation) products of genes are present even in functionally mature ejaculated sperm but are products of later spermatids (or earlier) active gene expression, processes that cease before spermiation. Recent analyses are challenging this belief suggesting that the rich repertoire of coding and non-coding RNAs in sperm is not a haphazard remnant from spermatogenesis in the testes but a carefully selectively retained and functionally coherent collection of RNAs (Das et al., Reference Das, McCarthy, Vishnoi, Paria, Gresham, Li, Kachroo, Sudderth, Teague, Love, Varner, Chowdhary and Raudsepp2013). More recent interpretations suggest human sperm retain mRNA that can be translated into protein in the oocyte after fertilisation (Jodar et al., Reference Jodar, Selvaraju, Sendler, Diamond and Krawetz2013). However, their precise role in the regulation of fertilisation and early embryonic development in the bovine remains to be determined. The mRNA expression of proteins associated with sperm function in bulls of high and low SCR reported a number of genes correlated with fertility status (Kasimanickam et al., Reference Kasimanickam, Kasimanickam, Arangasamy, Saberivand, Stevenson and Kastelic2012). Feugang et al. (Reference Feugang, Rodriguez-Osorio, Kaya, Wang, Page, Ostermeier, Topper and Memili2010) analysed the RNA profiles of sperm from high and low-fertility Holstein bulls using Affymetrix bovine genechips and reported differential expression in the abundance of mRNAs. A total of 415 transcripts out of ~24 000 were differentially detected in sperm collected from both fertility groups. Sperm from the low-fertility bulls were deficient of transcripts for transcriptional and translational factors while sperm from high fertility bulls contained higher concentrations of transcripts for extracellular space and membrane protein locations.
Short non-coding microRNAs (miRNAs) do not code for proteins, but various studies have reported that miRNAs regulate gene expression and also play a major role in embryo development (Boerke et al., Reference Boerke, Dieleman and Gadella2007). However, their precise role in the regulation of fertilisation and early embryonic development in the bovine remains to be determined. miRNA profiling from high and low-fertility bulls has also been previously performed (Govindaraju et al., Reference Govindaraju, Uzun, Robertson, Atli, Kaya, Topper, Crate, Padbury, Perkins and Memili2012) with seven miRNAs (aga-3155, -8197, -6727, -11796, -14189, -6125, -13659) being differentially expressed. Tscherner et al. (Reference Tscherner, Gilchrist, Smith, Blondin, Gillis and LaMarre2014) reported that the miR-34 miRNAs play a role in developing bovine gametes and suggested that individual variation in sperm miR-34 family abundance may be a biomarker of male bovine fertility. In addition, single nucleotide polymorphisms in target mRNA or miRNA have revealed associations with traits of economic interest and highlight the potential use of miRNAs in future genomic selection programs (Fatima and Morris, Reference Fatima and Morris2013). Sperm miRNA may be useful in understanding the transmission of epigenetic characteristics to male calves and its connection with the transgenerational inheritance of fertility/subfertility related traits. High-throughput RNA sequencing approaches will aid in the determination of the key coding and non-coding transcripts controlling sperm function and thus SCR.
Future directions of research directed at understanding the aetiology of idiopathic bull fertility
Male fertility has received far less attention in comparison to female fertility yet it is undoubtedly complex and definitely multifactorial. Despite many positive findings, the small numbers of bulls and, in some cases, an unreliable fertility phenotype due to insufficient insemination records for individual bulls as well as issues around sperm number used make interpretation of the findings of many studies challenging and sometimes unrepeatable when applied to different datasets. Despite this, it is now clear that that the sperm deliver not only DNA but also RNA and signalling factors to the oocyte at fertilisation. The most fruitful avenues of further investigation would appear to be around the differences among bulls in the kinetics of sperm penetration as well as completion of the first cell cycle and of the first mitotic cleavage after fertilisation. Embryos that cleave first are most likely to successfully reach the blastocyst stage and the quality of these embryos is superior at the preimplantation stage than later developing embryos. The pathophysiology of delayed cleavage may reside with the non-coding RNAs and or alterations in epigenetic signatures within the sperm which are most likely to be altered during testicular development or by epididymal modifications. An in-depth examination of these factors may shed new light on the cross-talk between bovine sperm and the early stages of embryo development; and importantly how this may be perturbed in bulls of low fertility. Future studies will no doubt take advantage of recent advances in high-throughput techniques to study DNA, RNAs, proteins, lipids, glycans and metabolites in combination. These ‘OMICS’-based technologies have increased our capacity to study new and novel aspects of sperm function and to get a broader view of these complex biological systems. They hold the main advantage of providing large volumes of information at relatively low cost and recent advances in bioinformatics enable the analysis and interpretation of large datasets in a more integrated systems biology approach.
Like so many studies thus far, these technologies will undoubtedly produce lists of biomarkers that are different between bulls of varying fertility. The major challenge then is to define which ones are physiologically important. For this, we need novel functional approaches comprising of both in vitro and in vivo methods. However, as outlined earlier in this review, before we go down this path we must be cognisant of the limitations of sire fertility estimates especially when inseminations are performed with high numbers of sperm. Then, we should ensure experiments are sufficiently powered with bulls across a wide range of the fertility spectrum in the quest to identify the reasons for the variation in SCR.
Acknowledgements
The authors acknowledge the contribution of Ms Edel Murphy in the compilation of this manuscript. The authors are supported by funding from Science Foundation Ireland (grant no. 16/IA/4474).
Declaration of interest
The authors have no vested interests to declare.
Ethics statement
This manuscript is a review of existing published work and as such did not use animals for its’ preparation.
Software and data repository resources
None.






