Introduction
Antimicrobial resistance (AMR) is a serious global public health challenge. Antibiotic resistance in human pathogens can cause treatment failure, prolong the duration of illnesses and increase mortality rates, exacting high human and economic costs to society (Friedman et al., Reference Friedman, Temkin and Carmeli2016). The wide and increasing use of antibiotics and other antimicrobial agents in human medicine, veterinary medicine, animal husbandry, horticulture and around the household have enhanced the selection and spread of antibiotic resistant bacteria (ARB) and antibiotic resistance genes (ARGs) (Baker-Austin et al., Reference Baker-Austin, Wright, Stepanauskas and Mcarthur2006; Meek et al., Reference Meek, Vyas and Piddock2015; O'Neill, Reference O'Neill2015).
The possible role of the natural environment, and surface water in particular, in transmission pathways of ARB and associated ARG has been the subject of much recent discussion (Wooldridge, Reference Wooldridge2012; Woolhouse et al., Reference Woolhouse, Ward, Van Bunnik and Farrar2015). A range of human activities, including activities of daily living, medical care and agriculture, generate waste that contains varying levels of antibiotics (and metabolites), ARB, and ARG. This waste is ultimately released into environmental media. Point sources, defined as ‘any single identifiable source of pollution from which pollutants are discharged’ (Armon and Starosvetsky, Reference Armon, Starosvetsky, Armon and Hänninen2015), represent an important and definable contribution to this effluent stream.
Once in the environment, these ARB and ARGs pose potential health risks to humans and animals (Ashbolt et al., Reference Ashbolt, Amezquita, Backhaus, Borriello, Brandt, Collignon, Coors, Finley, Gaze, Heberer, Lawrence, Larsson, Mcewen, Ryan, Schonfeld, Silley, Snape, Van Den Eede and Topp2013). They can persist in the environment, spread over land and water, and be transmitted via free-ranging wildlife (Baquero et al., Reference Baquero, Martinez and Canton2008; Berendonk et al., Reference Berendonk, Manaia, Merlin, Fatta-Kassinos, Cytryn, Walsh, Bürgmann, Sørum, Norström and Pons2015; Vittecoq et al., Reference Vittecoq, Godreuil, Prugnolle, Durand, Brazier, Renaud, Arnal, Aberkane, Jean-Pierre and Gauthier-Clerc2016). Within environmental niches, ARGs can increase clonally when a bacterial cell hosting an ARG divides, or be transferred between bacterial cells through horizontal gene transfer (HGT) (Allen et al., Reference Allen, Donato, Wang, Cloud-Hansen, Davies and Handelsman2010; Ashbolt et al., Reference Ashbolt, Amezquita, Backhaus, Borriello, Brandt, Collignon, Coors, Finley, Gaze, Heberer, Lawrence, Larsson, Mcewen, Ryan, Schonfeld, Silley, Snape, Van Den Eede and Topp2013; Berglund, Reference Berglund2015).
Despite an increase in the number of studies reporting AMR in diverse natural environmental media, including water, soil, sediment and wildlife, the relative contribution of specific anthropogenic sources to the quantity of ARB and ARGs in the environment is an area of debate (Wooldridge, Reference Wooldridge2012; Woolhouse et al., Reference Woolhouse, Ward, Van Bunnik and Farrar2015; Williams-Nguyen et al., Reference Williams-Nguyen, Sallach, Bartelt-Hunt, Boxall, Durso, Mclain, Singer, Snow and Zilles2016b). Therefore, in this study we sought to systematically identify and summarize evidence in the existing scientific literature pertaining to an association between effluent point sources and the quantity of ARGs in adjacent environmental media. In particular, we looked for measures of impact (i.e. effect measures), which quantify the magnitude or strength of the effect between a point source(s) and the frequency or concentration of resistance elements in the surrounding environment. The specific review question was: Is the prevalence or concentration of antibiotic resistance genes in soil, water, air or free-living wildlife higher in close proximity to, downstream from or downwind from, known or suspected sources compared to areas more distant, upstream, or upwind from these sources?
Because the majority of bacteria cannot be cultured, many researchers have begun to measure bacterial genes, including ARGs, in environmental media using culture-independent methods (Luby et al., Reference Luby, Ibekwe, Zilles and Pruden2016). These approaches, such as quantitative real-time polymerase chain reaction (q-PCR) and metagenomics (Henriques et al., Reference Henriques, Alves, Saavedra, Montforts and Correia2011), are able to provide insight into the environmental resistome in a way not possible using other technologies that rely on culture-dependent methods. Here we report systematic review results pertaining to ARG outcomes (ascertained via culture-independent methods).
Methods
The steps of the systematic review process are summarized in Fig. 1. A systematic review of the literature was conducted following a previously published protocol (Williams-Nguyen et al., Reference Williams-Nguyen, Bueno, Sargeant, Nault and Singer2016a) using the population, exposure, comparator, outcome, study design (PECOS) framework. The population of interest refers to environmental samples; soil, water, air, or free-living wild animal samples as such were considered. Non-wild animals were not considered as environmental samples because they are not a naturally occurring component of environmental systems free of humans.
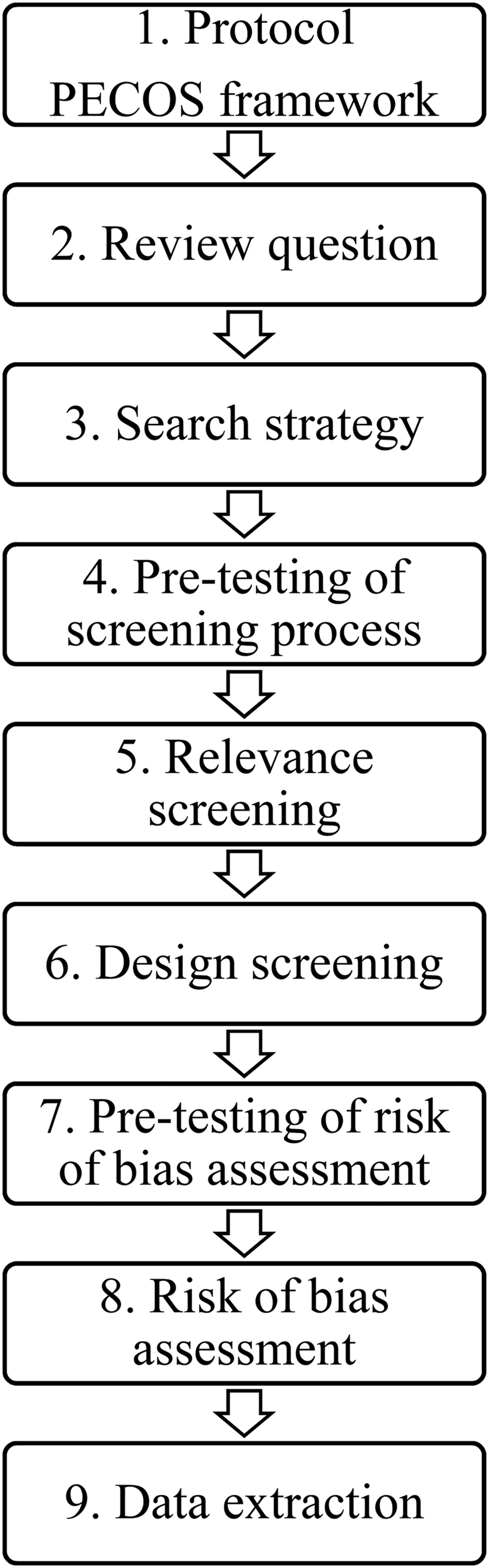
Fig. 1. Diagram summarizing the steps of the systematic review process.
The systematic review team was composed of six people, which included expertise on AMR, epidemiology, and systematic review methodology. PubMed©, Commonwealth Agricultural Bureaux (CAB Abstracts©), and Scopus© were searched on 14 October 2014 from inception date using specific search strategies. The search was updated on 19 April 2016 using identical search terms. The PubMed© controlled-vocabulary search string was as follows:
“drug resistance, microbial”[Mesh] AND (“water pollutants” [Mesh] OR “environment”[MeSH Terms] OR “soil”[MeSH Terms] OR “water”[MeSH Terms] OR “water pollution”[MeSH Terms] OR “air pollution”[MeSH Terms] OR “air pollutants” [MeSH Terms] OR “animals, wild”[MeSH Terms]) AND (“Animals”[MeSH Terms] OR “humans”[MeSH Terms] OR “animal feed”[MeSH Terms] OR “manure”[MeSH Terms] OR “aquaculture”[MeSH Terms] OR “waste water”[MeSH Terms] OR “sewage”[MeSH Terms] OR “hospitals”[MeSH Terms] OR “hospitals, animal”[MeSH Terms] OR “cities”[MeSH Terms]) NOT “therapeutics”[MeSH Terms] NOT “drug discovery” [MeSH Terms] NOT “aids”[All Fields] NOT “hiv”[All Fields] NOT “influenza”[All Fields].
The search string for CAB Abstracts© was:
(“Drug Resistance”.mp. and (“environment$” or “soil” or “water” or “water pollution” or “air pollut$” or “wild animals”).hw. and (“animals” or “man” or “feeds” or “manures” or “aquaculture” or “wastewater$” or “sewage” or “hospitals” or “animal hospitals” or “urban areas”).hw.) not “Therapeutics”.af. not “Drug discovery”.af. not “aids”.af. not “hiv”.af. not “influenza”.af.
The search string for Scopus© was:
TITLE-ABS-KEY ((antibiotic OR antimicrob*) AND resistan*) AND KEY (“environment*” OR “soil” OR “water” OR “water pollution” OR “air pollut*” OR “wild animals”) AND KEY (“animals” OR “man” OR “feeds” OR “manures” OR “aquaculture” OR “wastewater*” OR “sewage” OR “hospitals” OR “animal hospitals” OR “urban areas”) AND NOT TITLE-ABS-KEY (“Therapeutics”) AND NOT TITLE-ABS-KEY (“Drug discovery”) AND NOT TITLE-ABS-KEY (“aids”) AND NOT TITLE-ABS-KEY (“hiv”) AND NOT TITLE-ABS-KEY (“influenza”)
The same protocol was used for both culture-independent (ARG) and culture-dependent (ARB) outcomes, and thus studies with both outcome types were assessed as a whole up to the data extraction process, at which point ARG and ARB outcomes were independently evaluated. Therefore, although the focus of this publication is ARGs, the initial results include publication numbers relevant to ARB and ARGs.
There were no language or geographical limits on the search. All citations were imported into the EndNote reference management software package (Thomson Reuters, Philadelphia, PA), and duplicate records were removed.
Titles and abstracts of all citations were then screened to include only those relevant to the question. Specifically, studies were included if they: (a) were primary research; (b) collected environmental samples (soil, water, sediment, air, biological samples from wildlife); and (c) reported prevalence or concentration of ARG. An additional exclusion criterion – not stated in the original protocol (Williams-Nguyen et al., Reference Williams-Nguyen, Bueno, Sargeant, Nault and Singer2016a) – was added that asked: ‘Does the study use microbial source tracking techniques?’ Microbial source tracking techniques compare characteristics of fecal bacteria isolated from environmental sources with characteristics of fecal bacteria from known sources in an effort to identify the source of environmental isolates. Because these types of studies often fail to compare sites based on physical distance or direction from the source (e.g. (Edge and Hill, Reference Edge and Hill2005; Dickerson et al., Reference Dickerson, Hagedorn and Hassall2007; Mthembu et al., Reference Mthembu, Biyela, Djarova and Basson2010; Murugan et al., Reference Murugan, Prabhakaran, Al-Sohaibani and Sekar2012)), such studies do not provide evidence for this systematic review question. Any study that did not meet all these criteria was excluded. Those studies where it could not be ascertained from the title and abstract if they met all criteria were considered ‘unclear’ and passed through to the following screening phase for further clarification.
Full-text of remaining articles was retrieved, and the methods section only was reviewed. It was then determined whether the methodology used for each study was adequate to answer the systematic review question using the following inclusion criteria. Studies were included if they: (a) reported proximity to, or direction from a potential point source; and (b) had a comparison group (i.e. samples taken a fixed distance from or upstream from the source) or compared across a range of distances (i.e. samples taken at different distances from the source). Those studies that did not meet both criteria were excluded. An additional question not stated in the protocol (Williams-Nguyen et al., Reference Williams-Nguyen, Bueno, Sargeant, Nault and Singer2016a) a priori was added at this screening stage as follows: ‘Does the study implicitly or explicitly define a point source with reference to which a comparison was defined?’. During this screening phase, articles not written in English were identified, and an effort was made to translate the full text as review team resources allowed.
Pre-testing of the screening process was done by reviewing a sample of articles among all the citations from the database. Specifically, four articles that featured comparison groups based on information in the title or abstract were chosen. Papers of this type were selected to ensure testing of the second screening level (design screening). Two independent reviewers evaluated this phase, and improvements to the screening process and data entry were made based upon the reviewer's feedback. Final screening decisions were entered into a spreadsheet designed for this systematic review (Microsoft Office Excel 2013® Microsoft Corporation, Redmond, WA, USA).
For both screening phases (title/abstract and methods section of the article's full-text), two reviewers independently assessed each record. Consensus was required, and conflicts were resolved through phone conferences and e-mail.
Following the application of the inclusion/exclusion criteria, the full-text of each included study was evaluated for potential threats to internal validity (risk of bias assessment) by two independent reviewers per article. A customized relational database (Microsoft Access 2013®) was used for data entry on the risk of bias assessment. First, a qualitative rubric (explained below) was pre-tested by reviewing a sample from the included full-text articles after the two screening stages by three independent reviewers. A total of three articles were evaluated for this purpose. The pre-testing improved the consistency of the risk of bias assessment across reviewers, as well as the design of the data entry tool.
Articles were divided equally between each participating reviewer. A qualitative rubric of low, high, and unclear was assigned to each study for the potential risk of bias in the reported effect measure or other outcome variable due to selection bias, information bias, and confounding (Williams-Nguyen et al., Reference Williams-Nguyen, Bueno, Sargeant, Nault and Singer2016a). The risk of bias assessment was conducted at the study level and not at the outcome level due to the large number of possible outcomes per study. Selection bias was defined as systematic differences between the comparison groups with respect to how samples were collected in the study (methods used across sites). Information bias was defined as systematic differences in the methods for ascertaining ARG between comparison groups (i.e. use of different laboratory methods for the samples in the comparison groups). Confounding was evaluated with respect to the presence of point or non-point sources other than the source of interest that could have affected the study outcomes. It was assumed that a study that assessed the impact of a point source using sampling locations within a large spatial scale (e.g. 100 km distance between sampling locations) was at higher risk of confounding than a study where the spatial scale was smaller (e.g. a 10 km scale) due to the possible influence on the outcome of a larger number of alternative point and non-point sources, unless adequate confounding control measures were described. For all three types of biases, strategies to control or minimize the impact of these biases on the internal validity of the study were factored into the decision to classify them as low, unclear or high.
A final qualitative (low, high, and unclear) overall bias rubric was assigned to each study by considering the risk of bias from each domain after consensus was reached between the reviewers. In general, if a study had at least one out of the three domains classified as high risk, the overall result was considered high risk of bias, and the same applied for unclear risk of bias. However, there were exceptions, and the overall decision was made on a case-by-case basis relying on the judgment of the three reviewers involved in the risk of bias assessment.
Data from all studies, including the ones that were deemed to be at high risk of bias, were extracted and synthesized. Data consisted of characteristics of the study (geographic location, publication year, spatial scale, sampling design, type of laboratory detection method used), the exposure (point source), and the outcome: ARG prevalence or ARG concentration (either relative gene abundance, defined as ARG copies normalized to 16S copies, (2) absolute gene abundance or (3) gene concentration, defined as ARG copies divided by a measurement of volume), as reported by the authors, without further manipulation of that data. Any available information on statistical methods or modeling approaches used, and effect measures (and variability) reported for the comparison of interest were recorded. Data were entered into the same custom relational database, albeit in a different table from the one used for the risk of bias assessment. Additionally, a summary of the most relevant findings for the comparison of interest from each individual study was conducted and is presented in Tables 2 and 3.
In contrast to the original protocol (Williams-Nguyen et al., Reference Williams-Nguyen, Bueno, Sargeant, Nault and Singer2016a), the risk of bias assessment was conducted prior to the data extraction. To minimize introduction of bias by conducting these steps in reverse order, the reviewers who assessed studies during the risk of bias stage did not review the same studies during the data extraction, and were blinded to the risk of bias assessment decisions. Afterwards, a review team member uninvolved in either risk of bias assessment or data extraction validated all extracted data.
Results
The total number of records (including both culture-dependent to ascertain ARB and culture-independent methods for ARGs) returned by search strings totaled 5247 after de-duplication. The number of articles remaining after each screening step was 813 and 75, respectively. In total, 27 of the 75 included articles used culture-independent methods to ascertain ARGs. At the point of data extraction, three studies were identified wherein data were presented as aggregated and no qualitative or quantitative comparison of ARG prevalence or ARG concentration by distance or direction from the source was available. Therefore, these studies were excluded as providing no information about this systematic review question (Auerbach et al., Reference Auerbach, Seyfried and Mcmahon2007; Bajaj et al., Reference Bajaj, Singh, Kanaujia and Virdi2015; Xi et al., Reference Xi, Zhang, Kwok, Huo, Feng, Zhang and Sun2015). Hence, the final number of studies assessed in this review was 24 (Fig. 2).
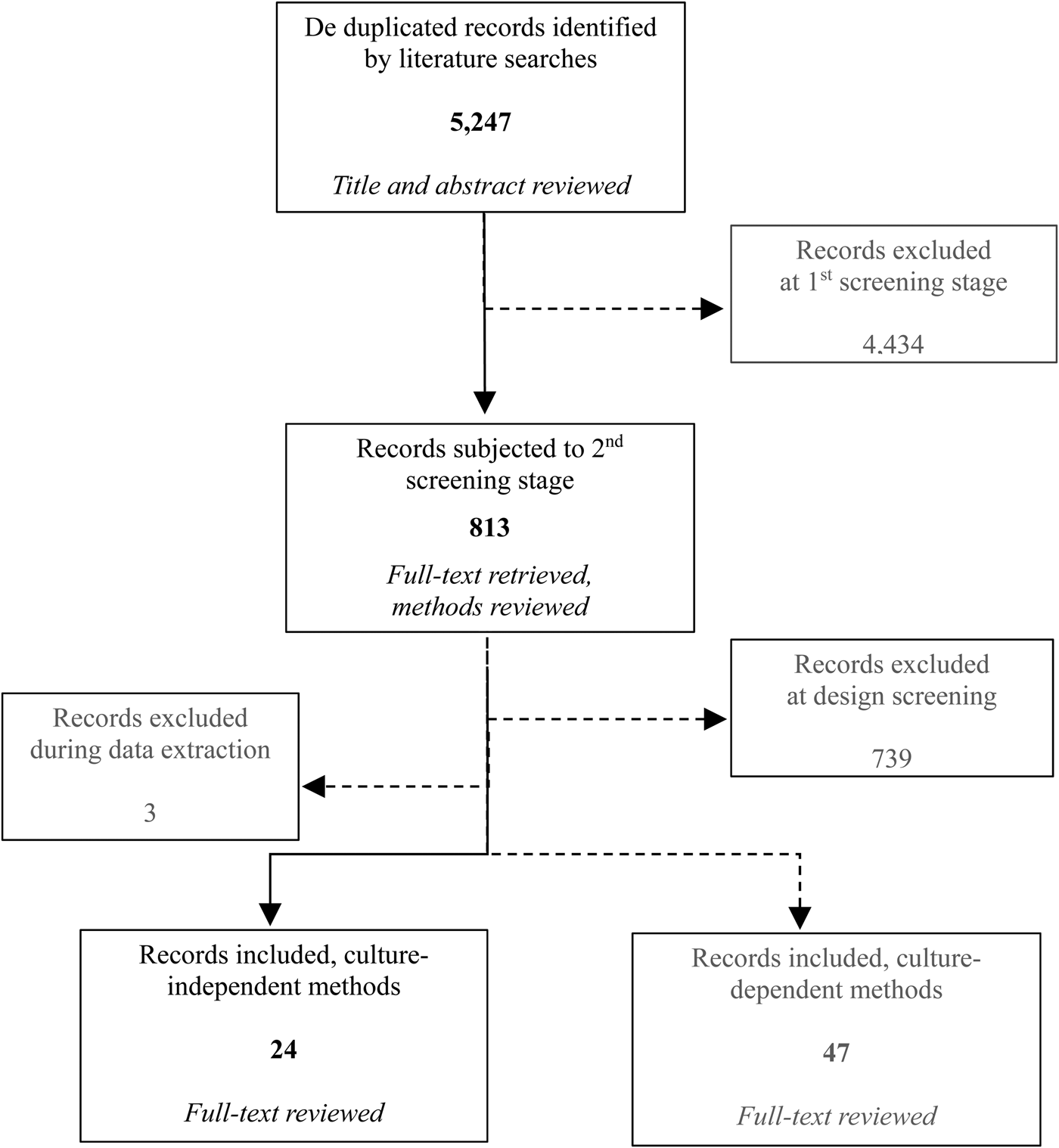
Fig. 2. Flowchart summarizing the selection process for the studies (the shaded boxes depict the articles excluded from the process and the records for the ARB outcome, not assessed in this paper).
For the overall risk of bias assessment, three studies were categorized as high risk of bias, 13 were at an unclear risk of bias, and eight were deemed to be at low risk for bias. The rubric for the risk of bias levels was previously published (Williams-Nguyen et al., Reference Williams-Nguyen, Bueno, Sargeant, Nault and Singer2016a).
An example of a study considered at high risk of bias was Zhang et al. (Reference Zhang, Lv, Zhang, Zhang and Yu2013). This study involved collection of samples at a spatial scale of about 50 km and did not adjust for potential confounders in the analysis, such as other point or non-point sources in the 50 km study area. An example of a low risk of bias study was Pruden et al. (Reference Pruden, Arabi and Storteboom2012). Despite a spatial scale of more than 100 km, this study controlled for potential confounders from many other sources of anthropogenic effluent by using linear regression modeling to account for distance to different source types. An example of a study with unclear risk of bias was Lapara et al. (Reference Lapara, Burch, Mcnamara, Tan, Yan and Eichmiller2011). In this study, description on the selection of samples at different locations was lacking. Additionally, there were possible confounders such as effluent from non-point sources from agricultural and recreational water use that were not mentioned. Given the lack of information, it was not possible to determine if the risk of bias of estimates of the relationship between the source of interest (WWTP) and ARG concentration (intl1, tetA, tetX, and tetW) in river and lake surface waters and sediments was high or low in this study, and thus it was classified as unclear.
While all included studies were written in English, one study written in Chinese (Liu et al., Reference Liu, Mao, Ren, Luo and Cao2012) was deemed relevant to the review question based on the title and abstract that were available in English; however, full-text translation was not feasible, hence it is uncertain if it would have been finally included. No other non-English articles met eligibility criteria based on English title or abstract (or translations of the title/abstract).
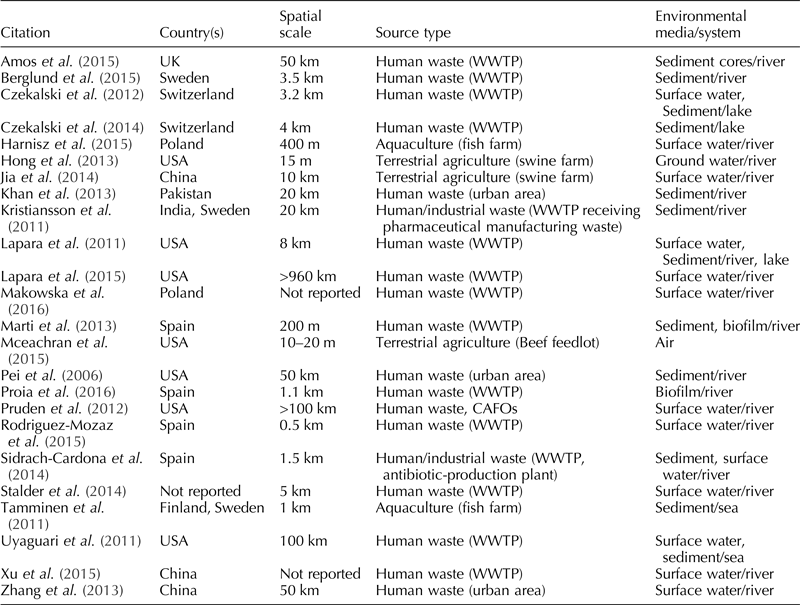
The geographic location of the studies (n = 24) was diverse: China (n = 3), Finland (n = 1), India (n = 1), Pakistan (n = 1), Poland (n = 2), Spain (n = 4), Sweden (n = 3), Switzerland (n = 2), UK (n = 1), USA (n = 7). There was one study (Stalder et al., Reference Stalder, Barraud, Jové, Casellas, Gaschet, Dagot and Ploy2014) in which the location could not be ascertained after reviewing the full-text, and two studies involved two different countries (Kristiansson et al., Reference Kristiansson, Fick, Janzon, Grabic, Rutgersson, Weijdegard, Soderstrom and Larsson2011; Tamminen et al., Reference Tamminen, Karkman, Lohmus, Muziasari, Takasu, Wada, Suzuki and Virta2011).
Date of publication ranged from 2006 to 2016, with the highest number of publications in 2015 (n = 7). The spatial scale for the sampling frame ranged from 10–20 m (McEachran et al., Reference McEachran, Blackwell, Hanson, Wooten, Mayer, Cox and Smith2015) to more than 900 km (Lapara et al., Reference Lapara, Madson, Borchardt, Lang and Johnson2015).
The majority of studies investigated not only point sources of human waste, especially wastewater treatment plants (n = 16), but also urban areas (n = 3). Terrestrial animal agriculture was examined in three studies: two studies examined swine farms and one study examined a beef cattle feedlot. Aquaculture (fish farms) was assessed in two studies.
Surface water was the most common type of environmental media sampled (n = 13), followed by sediment (n = 12), biofilm (n = 2), air (n = 1), and groundwater (n = 1). None of the included studies sampled wildlife. Five of the studies collected more than one sample type. For a summary of the sampling information, see Table 1.
Table 1. Descriptive information for each one of the 24 studies included in this systematic review
Overall, the most common target gene outcomes were sul1 (n = 12), tetW (n = 11), tetA (n = 9), and sul2 (n = 8). The number of genes per study ranged from 1 to 13, with the majority of studies evaluating four different genes (n = 7). Most studies used q-PCR to ascertain ARGs (n = 23), and one study used shotgun metagenomics (Kristiansson et al., Reference Kristiansson, Fick, Janzon, Grabic, Rutgersson, Weijdegard, Soderstrom and Larsson2011).
Regarding the outcome data type, 20 studies compared relative gene abundance only, three compared absolute gene concentration only, and one study compared both relative gene abundance and absolute gene concentration. None of the studies used prevalence as their outcome type.
With reference to statistical methods and modeling approaches, nine out of the 24 studies conducted statistical analysis to compare ARG outcomes upstream vs. downstream (or near vs. far sites) with reference to a single point source, and three out of the 24 studies used modeling approaches to describe the effect of multiple sources. However, no effect measures were described in any study. Specifically, one study used a t-test to compare relative gene abundance of each one of the target ARG between upstream and downstream sites from a WWTP (Berglund et al., Reference Berglund, Fick and Lindgren2015). Eight studies compared the relative gene abundance (Khan et al., Reference Khan, Berglund, Khan, Lindgren and Fick2013; Marti et al., Reference Marti, Jofre and Balcazar2013; Stalder et al., Reference Stalder, Barraud, Jové, Casellas, Gaschet, Dagot and Ploy2014; Harnisz et al., Reference Harnisz, Korzeniewska and Gołaś2015; Makowska et al., Reference Makowska, Koczura and Mokracka2016; Proia et al., Reference Proia, Von Schiller, Sànchez-Melsió, Sabater, Borrego, Rodríguez-Mozaz and Balcázar2016) or the absolute gene concentration (Uyaguari et al., Reference Uyaguari, Fichot, Scott and Norman2011; Rodriguez-Mozaz et al., Reference Rodriguez-Mozaz, Chamorro, Marti, Huerta, Gros, Sànchez-Melsió, Borrego, Barceló and Balcázar2015) across sites using either ANOVA or a non-parametric method for comparison of means such as Kruskal–Wallis, Friedman, or Mann–Whitney tests at the 0.05 significance level. One study compared the relative gene abundance of ARGs across sites based on distance from the source using graphical regression and interpolation (Czekalski et al., Reference Czekalski, Gascon Diez and Burgmann2014). Of the nine studies that reported statistical inference, six found a significant relationship for the majority of the target ARG (Uyaguari et al., Reference Uyaguari, Fichot, Scott and Norman2011; Khan et al., Reference Khan, Berglund, Khan, Lindgren and Fick2013; Marti et al., Reference Marti, Jofre and Balcazar2013; Berglund et al., Reference Berglund, Fick and Lindgren2015; McEachran et al., Reference McEachran, Blackwell, Hanson, Wooten, Mayer, Cox and Smith2015; Proia et al., Reference Proia, Von Schiller, Sànchez-Melsió, Sabater, Borrego, Rodríguez-Mozaz and Balcázar2016), and three did not (Stalder et al., Reference Stalder, Barraud, Jové, Casellas, Gaschet, Dagot and Ploy2014; Harnisz et al., Reference Harnisz, Korzeniewska and Gołaś2015; Makowska et al., Reference Makowska, Koczura and Mokracka2016). Of the three studies that conducted modeling approaches, Amos et al. (Reference Amos, Gozzard, Carter, Mead, Bowes, Hawkey, Zhang, Singer, Gaze and Wellington2015) used a log–log regression model to explain the relative abundance of intl1 in river sediment samples at sites across a range of WWTP outputs, adjusting for other variables; Lapara et al. (Reference Lapara, Madson, Borchardt, Lang and Johnson2015) used a fluid-kinetics (plug-flow) model to explain the relative abundance of ARG in a river as a function of several variables, including distance from the multiple WWTP; and Pruden et al. (Reference Pruden, Arabi and Storteboom2012) conducted general linear regression models to explain the log relative gene abundance along a river with an exposure gradient as a function of several variables.
In the section that follows, results are summarized for each group of point source investigated (human waste and animal agriculture) by the type of comparison made (upstream vs. downstream in rivers or based on distance from the source) and by type of outcome reported (relative gene abundance or absolute gene concentration).
Human waste (n = 19)
From the 19 studies, 16 assessed WWTP and/or industrial waste, and three studies urban areas. Among the 16 that evaluated WWTP and/or industrial waste, 13 compared ARG outcomes in unidirectional systems (n = 10) or based on distance (n = 3) with reference to a single point source, while three studies described the effect of multiple point sources using modeling approaches.
Among the 10 studies that assessed the impact of WWTP and/or industrial waste in unidirectional systems (i.e. rivers), eight reported relative gene abundance only, one reported absolute gene concentration only, and one reported both. Among the first group, four studies showed a higher relative gene abundance at downstream sites from the source compared with upstream sites (Kristiansson et al., Reference Kristiansson, Fick, Janzon, Grabic, Rutgersson, Weijdegard, Soderstrom and Larsson2011; Berglund et al., Reference Berglund, Fick and Lindgren2015; Makowska et al., Reference Makowska, Koczura and Mokracka2016; Proia et al., Reference Proia, Von Schiller, Sànchez-Melsió, Sabater, Borrego, Rodríguez-Mozaz and Balcázar2016). One study reported no difference in relative gene abundance downstream compared with upstream (Stalder et al., Reference Stalder, Barraud, Jové, Casellas, Gaschet, Dagot and Ploy2014). The remaining three studies presented conflicting evidence for the effect of WWTP/industrial waste on the relative gene abundance (Marti et al., Reference Marti, Jofre and Balcazar2013; Sidrach-Cardona et al., Reference Sidrach-Cardona, Hijosa-Valsero, Marti, Balcázar and Becares2014; Xu et al., Reference Xu, Xu, Wang, Guo, Qiu, He, Zhang, Li and Meng2015). The only study that evaluated absolute gene concentration presented conflicting evidence (Rodriguez-Mozaz et al., Reference Rodriguez-Mozaz, Chamorro, Marti, Huerta, Gros, Sànchez-Melsió, Borrego, Barceló and Balcázar2015). Finally, Uyaguari et al. (Reference Uyaguari, Fichot, Scott and Norman2011) reported both a lower gene concentration and a lower relative gene abundance downstream.
Three studies assessed the impact of WWTP across a range of distances. Two of them reported relative gene abundance and one study reported gene concentration. Among the former group, one study found higher relative gene abundance at sites closer to the source compared with distant sites (Czekalski et al., Reference Czekalski, Gascon Diez and Burgmann2014) and the other study found no difference in relative gene abundance between near and far sites (Czekalski et al., Reference Czekalski, Berthold, Caucci, Egli and Bürgmann2012). The study reporting gene concentration found higher gene concentration at sites closer to the source compared with distant sites (Lapara et al., Reference Lapara, Burch, Mcnamara, Tan, Yan and Eichmiller2011). The remaining three studies assessing the impact of WWTP conducted modeling approaches and they all reported relative gene abundance. The model conducted by Amos et al. (Reference Amos, Gozzard, Carter, Mead, Bowes, Hawkey, Zhang, Singer, Gaze and Wellington2015) indicated that a 10% increase in the total WWTP impact (defined as a function of type of, size of, and river course distance from upstream WWTPs) at a given site was associated with a 3.2% increase in the relative abundance of intl1, adjusting for land cover, season, and rainfall. The fluid kinetics model predictions by Lapara et al. (Reference Lapara, Madson, Borchardt, Lang and Johnson2015) for the Mississippi river did not show a good fit for the target genes, and the general linear regression models in Pruden et al. (Reference Pruden, Arabi and Storteboom2012) in a river system in Colorado showed an association between average log relative sul1 abundance and the impact of inverse-distance weighted upstream WWTP and animal feeding operation capacities; however, they did not find such an association for the other target gene (tetW).
The three studies that assessed the impact of urban areas as the source of human waste reported relative gene abundance in river systems. Khan et al. (Reference Khan, Berglund, Khan, Lindgren and Fick2013) found a higher relative gene abundance downstream compared with upstream sites; Zhang et al. (Reference Zhang, Lv, Zhang, Zhang and Yu2013) found no difference between upstream and downstream sites from a city; and Pei et al. (Reference Pei, Kim, Carlson and Pruden2006) reported mixed evidence depending on the sampling season (high vs. low water flow) and on the target genes.
Animal agriculture (n = 5)
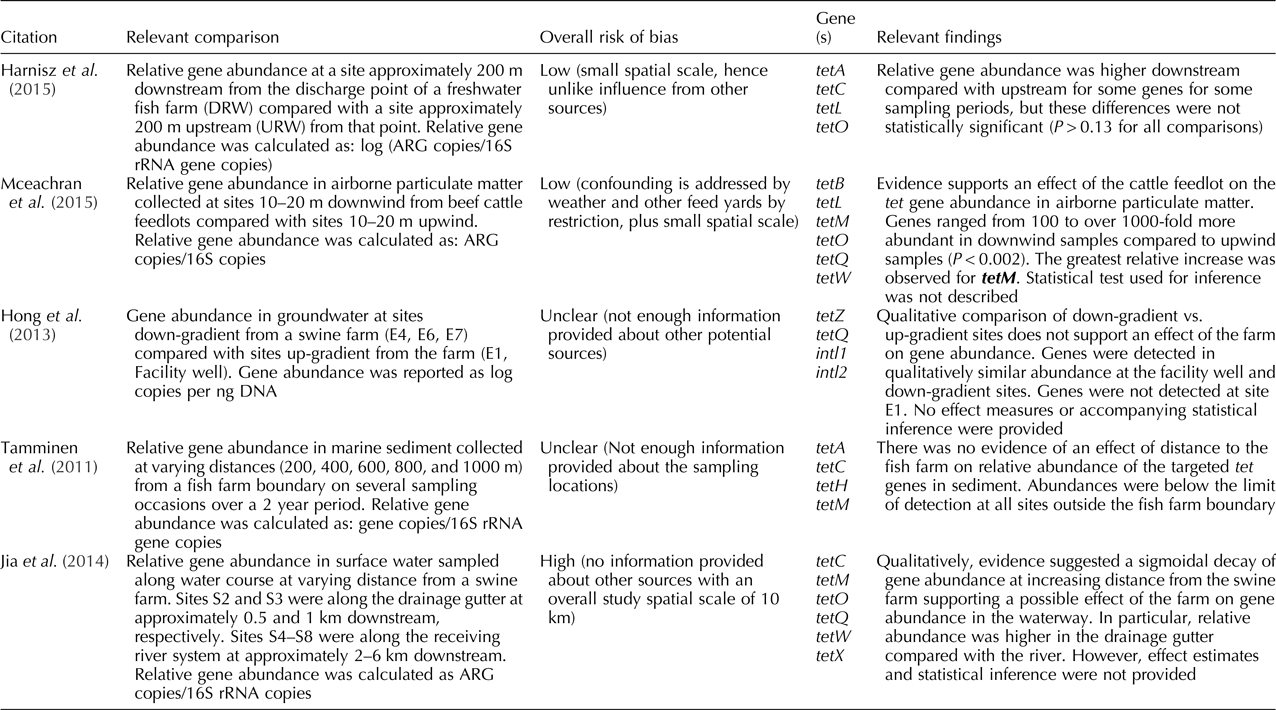
Of these five studies, three assessed terrestrial agriculture and two aquaculture. Among the three studies that assessed the impact of terrestrial animal agriculture, two were conducted in unidirectional systems (i.e. rivers), of which one reported relative gene abundance, and one absolute gene abundance. Specifically, McEachran et al. (Reference McEachran, Blackwell, Hanson, Wooten, Mayer, Cox and Smith2015) reported a higher relative gene abundance downwind compared with upwind sites from beef cattle feedlots; and Hong et al. (Reference Hong, Yannarell, Dai, Ekizoglu and Mackie2013) did not find a difference in absolute gene abundance in groundwater samples between up-gradient and down-gradient sites from a swine farm. The third study examining terrestrial animal agriculture made comparisons based on distance from a swine farm, reporting higher relative gene abundance near the farm compared with sites farther away (Jia et al., Reference Jia, He, Bu, Shi, Miao, Zhou, Shan and Zhang2014).
The two studies assessing aquaculture as the point source made comparisons of relative gene abundance in a river system (Harnisz et al., Reference Harnisz, Korzeniewska and Gołaś2015) and based on distance from the fish farm (Tamminen et al., Reference Tamminen, Karkman, Lohmus, Muziasari, Takasu, Wada, Suzuki and Virta2011). Harnisz et al. (Reference Harnisz, Korzeniewska and Gołaś2015) found a higher relative abundance of some target genes downstream compared with upstream sites from a fish farm depending on the sampling season, while Tamminen et al. (Reference Tamminen, Karkman, Lohmus, Muziasari, Takasu, Wada, Suzuki and Virta2011) did not find an apparent impact of the fish farm across a range of distances on the relative gene abundance. For more details on the results for individual studies refer to Tables 2 and 3.
Table 2. Findings for the studies included in this systematic review that assessed human waste (WWTP, industrial, urban areas) as a point source, organized by risk of bias (from low to high) (n = 19).

In the Relevant findings column, specific genes from each study are emphasized in bold. WWTP: wastewater treatment plant.
Table 3. Findings for the studies included in the systematic review that had animal agriculture (both terrestrial and aquaculture) as a point source, presented by risk of bias (from low to high) (n = 5)
Discussion
This systematic review aimed to identify and summarize the available evidence on the impact of anthropogenic point sources on the increase of ARGs in the environment. Based on the authors’ prior knowledge of the literature on this subject, the assumption was made that etiologic research on this review question would be uncommon, and that a narrowly focused question would not provide sufficient evidence to be meaningfully summarized. Thus, the review question was broadly formulated, permitting evaluation of a larger pool of evidence but also increasing the heterogeneity among the studies.
Most studies were considered to be unclear for risk of bias. The common reason for this was lack of information about potential confounders that might bias the observed relationship between proximity to a point source and levels of ARG in environmental media. The predominant confounder of concern to the review team was the introduction of antibiotics, ARB, or ARGs from other sources that could differentially affect the exposed and comparator sites. Many studies did not provide details about other possible contributors to resistance in the system or did not explain the location of other contributors and sampling sites. Studies with moderate to large spatial scales and no information about potential confounders were common. Most of these were considered to be an unclear or high risk of confounding bias.
We note that risk of bias assessment was conducted before the data extraction, which is a deviation from the original protocol. Though non-standard, this is unlikely to have introduced additional biases into the review findings because different reviewers evaluated the same study at the two different stages, and we extracted data from all studies including those considered at high risk of bias. The risk of bias assessment was conducted using subjective judgment, and despite reviewer consensus, this is a limitation of this review process.
As we noted, the most commonly evaluated point source was WWTPs, which has been recognized to contain a large diversity of ARB and ARGs (Rizzo et al., Reference Rizzo, Manaia, Merlin, Schwartz, Dagot, Ploy, Michael and Fatta-Kassinos2013). Human waste, which can include antibiotics, bacteria, and potentially ARB and ARGs, is treated at WWTPs. However, ARGs are still found after the treatment process, at the WWTP discharge, or at sites downstream from the WWTP (Rizzo et al., Reference Rizzo, Manaia, Merlin, Schwartz, Dagot, Ploy, Michael and Fatta-Kassinos2013); most studies reported the highest levels of ARGs (relative gene abundance or concentration) in river sites downstream from the point source (the WWTP) as compared with upstream sites or near the WWTP (compared with sites far from it). Only five studies assessing the impact of animal agriculture (three terrestrial representing swine farms and a beef feedlot, and two in aquaculture representing fish farms) were included in the final pool of studies to review and reported mixed findings. The small number of studies, the heterogeneity among the animal systems and other gathered evidence revealed insufficient scientific evidence about the impact of animal agriculture on a measurable increase of antibiotic resistance in the surrounding environment. One potential reason for this knowledge gap is that agricultural farms are more challenging point sources to assess compared with wastewater treatment plants, highlighting the need for additional studies of animal systems.
Overall, there was consistency in the results for the outcome data types (relative or absolute gene abundance or gene concentration) with most studies reporting a higher relative gene abundance and /or gene concentration downstream from the source (in unidirectional studies) or near the source (for those studies based on distance) across all source types.
Across all studies in the review, sul1, sul2, tetA, and intl1 were the most frequently studied and detected ARGs. Sul1 and sul2, mainly found in Gram-negative bacteria (Sköld, Reference Sköld2000), confer resistance to sulfonamide antibiotics, which are used in both animal and human practice, by modifying the dihydropteroate synthase related to protein synthesis. Among the large group of tet genes, tetA confers resistance to tetracycline via efflux pumps (Roberts, Reference Roberts2005). In the case of intl1, it codes for the integrase enzyme associated with many drug resistant bacteria (Mazel, Reference Mazel2006). However, intl1, intl2, intl3 (integrons) are not always associated with AMR. Other ARG commonly detected were bla TEM, bla SHV, and ermB. The first two confer resistance to β-lactam antibiotics (e.g. penicillins, cephalosporins, carbapenems) by encoding for β-lactamase enzymes, and ermB confers resistance to macrolide antibiotics through the modification of 23S by rRNA methylation (Szczepanowski et al., Reference Szczepanowski, Linke, Krahn, Gartemann, Gutzkow, Eichler, Puhler and Schluter2009). None of the studies reported detecting floR (perhaps because they did not search for it specifically), which has a wide global distribution and has been found associated with both agriculture and aquaculture (Cloeckaert et al., Reference Cloeckaert, Baucheron, Flaujac, Schwarz, Kehrenberg, Martel and Chaslus-Dancla2000; Fernandez-Alarcon et al., Reference Fernandez-Alarcon, Miranda, Singer, Lopez, Rojas, Bello, Dominguez and Gonzalez-Rocha2010).
Among those studies that conducted a statistical analysis, ANOVA or an equivalent non-parametric method was the most common approach. Such methods for comparison of means (unlike regression methods) cannot produce a quantitative summary effect measure when used to evaluate complex systems with a large number of relevant comparison groups or covariates. A combination of regression methods such as the ones proposed by Pruden et al. (Reference Pruden, Arabi and Storteboom2012) and Amos et al. (Reference Amos, Gozzard, Carter, Mead, Bowes, Hawkey, Zhang, Singer, Gaze and Wellington2015) together with spatial analysis, as used in the study by Czekalski et al. (Reference Czekalski, Gascon Diez and Burgmann2014) can provide a good framework to address some of the challenges related to bias, and quantification of the impact of point sources on the prevalence or concentration of ARG in the environment.
In light of this review process, the protocol (Williams-Nguyen et al., Reference Williams-Nguyen, Bueno, Sargeant, Nault and Singer2016a) would have benefited from a few modifications (besides the ones we made a posteriori) to minimize the limitations and challenges encountered throughout the review. For instance, it would have been valuable to have an available tool to address the quality of the methodology and evidence provided by the studies for our specific review question; a possible solution to this would have been to include only those studies that explicitly defined a comparison of relevance to the review question or that conducted a statistical analysis for such a comparison.
Potential publication bias could not be assessed for this body of evidence. Publication bias is the exaggeration of treatment effect sizes caused by the propensity for journals to preferentially publish research showing statistically significant results (Song et al., Reference Song, Hooper and Loke2013). Such bias can cause a meta-analysis to give a misleading picture of the effect size in question, such that the average effect size appears to exist when none is truly present or to exaggerate the magnitude of a significant effect size. Quantitative assessment of the presence of publications bias is possible when the distribution of sufficiently homogeneous effect measures can be examined via funnel plots and other methods (Duval and Tweedie, Reference Duval and Tweedie2000). This review did not identify such a pool of quantitative results, thus publication bias could not be evaluated. Additionally, some existing evidence may not have been identified by our search. Although some of the databases searched do index grey literature, our search strategy did not identify any. Furthermore, a meta-analysis was not conducted in this review for the same reason (lack of quantifiable homogeneous outcomes).
We identified a number of important considerations for future studies seeking to estimate the effect of a specific point source on environmental levels of ARGs. Our review highlighted the need for epidemiological and/or ecological observational studies that control for selection bias, information bias, and confounding to the extent possible. Such studies will need to describe and adjust for confounders (especially due to other sources of antibiotics or resistant bacteria and/or genes). A good example of such an approach is the study by Pruden et al. (Reference Pruden, Arabi and Storteboom2012). Additionally, there remains a need for studies where the data analysis provides effect measures such as odds ratios or risk ratios (for studies with ARG prevalence as the outcome data type) or mean differences (for studies with ARG concentration as the outcome data type) to quantify the magnitude or strength of the effect of the exposure (i.e. the point source) on the outcome (i.e. the prevalence or concentration of ARG in the surrounding environment), accompanied by measures of variability. Pruden et al. (Reference Pruden, Arabi and Storteboom2012), despite creating relevant generalized linear regression models of the relationship of interest, did not provide parameter estimates from these models which would be needed to quantify the effect WWTPs had on ARGs after accounting for other sources such as animal feeding operations, and conversely, the effect that animal feeding operations on ARGs after accounting for WWTPs.
Similarly, researchers should use statistical methods to infer the significance of the study findings, and report these along with study results. The most appropriate statistical model(s) will depend on specifics of the study design and on the outcome of interest. Enhanced collaborative work between microbiologists, ecologists, and other scientists to provide expertise where needed will aid in successful efforts to conduct etiologic research.
There is no doubt that the increase of ARB is a global health crisis, and that there is a need to understand and to intervene with the dissemination pathways. The role of the natural environment in the dynamics of antibiotic resistance is an area of great interest and concern (Singer et al., Reference Singer, Ward and Maldonado2006; Allen et al., Reference Allen, Donato, Wang, Cloud-Hansen, Davies and Handelsman2010). Research on the issue must use methodology able to contend with the inherent complexity of environmental systems subject to flux, as well as the necessarily observational nature of most scientific evidence.
This systematic review provides a strong imperative to improve research methods in order to provide interpretable, quantitative information about the effect of point sources on resistance in the environment. Such information will ultimately be vital for developing effective interventions that will address resistance in the environment and benefit human and animal health.
Acknowledgments
We thank Dr Annette O'Connor (Iowa State University) for her helpful insight during the protocol development process.
Funding sources
Partial funding for this project was provided by the National Pork Board, United States (NPB Project #13-260). This funding source was not involved in any aspect of the review design, inclusion/exclusion decisions, data collection, or formulation of conclusions.








