Appendix: Methods for Mouse Patent Landscape and Impact Analysis
We searched the Thomson database Delphion on September 27, 2007, for granted US patents using the following search strategy. First we used a modification of the Ade/Cook-Deegan algorithm.Footnote 11 The algorithm restricts the search to relevant patent classes and searches claims for terms commonly associated with DNA/RNA patents: ((((((119* OR 426* OR 435* OR 514* OR 536022* OR 5360231 OR 536024* OR 536025* OR 800*) <in> NC)
AND ((antisense OR <case><wildcard>cDNA* OR centromere OR deoxyoligonucleotide OR deoxyribonucleic OR deoxyribonucleotide OR <case><wildcard>DNA* OR exon OR “gene” OR “genes” OR genetic OR genome OR genomic OR genotype OR haplotype OR intron OR <case><wildcard>mtDNA* OR nucleic OR nucleotide OR oligonucleotide OR oligodeoxynucleotide OR oligoribonucleotide OR plasmid OR polymorphism OR polynucleotide OR polyribonucleotide OR ribonucleotide OR ribonucleic OR “recombinant DNA” OR <case><wildcard>RNA* OR <case><wildcard>mRNA* OR <case><wildcard>rRNA* OR <case><wildcard>siRNA* OR <case><wildcard>snRNA* OR <case><wildcard>tRNA* OR ribonucleoprotein OR <case><wildcard>hnRNP* OR <case><wildcard>snRNP* OR <case><wildcard>SNP*) <in> CLAIMS))
AND (((mouse) OR (mus*) OR (mammal*) OR (musculus) OR (murine) OR (mice) OR (Mus musculus)))))
AND (((mammal*) <in> CLAIMS) OR ((mouse) <in> CLAIMS) OR ((mus*) <in> CLAIMS) OR ((murine) <in> CLAIMS) OR ((mice) <in> CLAIMS) OR ((musculus) <in> CLAIMS) OR ((Mus musculus) <in> CLAIMS)))
We then searched plant in claims (((Plant*) <in> CLAIMS)) and removed all patents from search one that were also found in search two.
We downloaded all available data fields for the 7179 candidate granted patent identified by our search, including title, publication date, original national class, publication number, publication country, number of claims, assignee/applicant name, assignee/applicant state/city, assignee/applicant country, USPTO assignee code, USPTO assignee name, application number, application date, application country, attorney name, domestic references, number of domestic references, forward references, number of forward references, foreign references, other references, designated states national, designated states regional, ECLA codes, Examiner – primary, Examiner – assistant, family patent numbers, inventor name, inventor city/state, inventor country, IPC-R codes, inventive IPC-R, IPC-7 codes, Main IPC-7, National class, Main national class, field of search, maintenance status code, number of pages, priority number, priority date, and priority country.
Patent Coding
We read and coded all claims of all 7179 patents to (1) identify those patents that potentially claim mouse gene sequences; (2) identify the SEQ IDs of gene sequences actually claimed by patents; and (3) add additional codes, including: the assignee type (public/private university, government agency, pharmaceutical or biotechnology company, nongovernmental organization and individual inventor), any methods claimed, cell type(s) claimed, or transgenic animals claimed.
Included in our final analysis were 1144 patents that claimed mouse genes, mostly in the form of nucleotide sequences, but also amino acid sequences and a small number that claimed a gene by name. Prior to 1996, US patents did not require the genetic sequences to be listed with an associated SEQ ID.
List of Patented Mouse Gene Sequences
The resulting list of patent number–sequence ID pairs was matched, using a simple Python script written by postdoctoral fellow at the University of Alberta, Dr. Andreas Strotmann, against the Cambia Patent Lens database of genetic sequences extracted from US patents retrieved in June 2008.Footnote 12
We retrieved, in FASTA format, nucleotide sequences for 32,351 DNA SEQ IDs in 929 patents and 179 amino acid SEQ IDs in 105 patents for a total of 32,530 sequences or sequence patterns listed in 983 patents (note that some patents listed both nucleotide and amino acid SEQ IDs). This data set was then manually filtered to retain only those sequences that were actually claimed in patents. We collected patented sequences that were not matched to the Patent Lens database from the Entrez database (if only the gene name was specified), from the patent claims themselves, or from the Patent Analysis website.Footnote 13
Determining Patented Mouse Genes
To determine the parts of the mouse genome that corresponded to the sequences in these patents, Dr. Songyan Liu, a bioinformatician, and his colleagues at the University of Manitoba performed a BLAST (basic local alignment search tool) analysis of all nucleotide and amino acid sequences identified earlier, using standard settings except for the following: Tag length ≥ 25; Expect < 0.001; Score ≥ 48 (Figure 10.A1). The Expect value setting means that there is a less than 1 in 1000 chance that the gene match is the result of pure chance. This is significantly lower than in the usual bioinformatics setting but higher than the Expect=0 exact match requirement in Murray and Jensen (2005).Footnote 14 The reasons for this choice are (1) most patent documents specifically state that they cover any genetic sequence similar to the one listed in the patent and (2) the sequence being patented and the corresponding sequence in the Ensembl database may be from different alleles of the same gene. In all cases, we retained only the best hit.

Figure 10.A1 Schematic of BLAST analysis – to match sequences identified from patents to the annotated mouse genome.
Using this method, we identified 1886 nucleotide sequences against the known mouse genome. An additional 62 entire genes were claimed by name or description rather than sequence. For the genes claimed by name or description, we searched the NCBI Entrez Gene database for entries matching their identifying description found in the patent claims. The resulting matches were added to the data set.
Our matching method identified 1692 genetic sequences from 952 mouse genes claimed, as a whole or in part, in 1049 US patent applications; including one mitochondrial gene (out of 37 known). This equates to 2.9% of the 32,480 mouse genes available in NCBI Mouse Build 37 against which we matched our sequences. Other sequences were from unknown species with low homology to the mouse genome, were for noncoding fragments (i.e., did not map onto known mouse genes) or were artificial sequences.
Collecting Information on Patented Mouse Genes
For each of the 952 identified genes, Dr. Songyan Liu, at the University of Manitoba, and his colleagues extracted the following information from bioinformatics databases in December 2008:
Trap hit: how many known hits were available for this gene.
Gene targeting status: 822 of the patented genes (86%) had a corresponding targeting request at one of the knockout mouse consortia.
OMIM information on the gene: 616 patented genes (65%) had an OMIM ID, 191 (20%) an OMIM description.Footnote 15
OMIM disease descriptors for 952 – (649 + 6) = 297 patented genes (31%).
MGI phenotypes available for each gene: 485 of the patented genes had some kind of phenotype listed (51%).Footnote 16
Detailed Gene Ontology information per gene – all functions, processes, and components where this gene is known to play a role; 888 of the genes (93%) had one or more gene ontology entries.
945 genes (99.2%) had entries for all three gene ontology components in the MGI Gene Ontology Slim Chart;Footnote 17 that is, seven of the patented genes were still classified as “novel genes” at MGI at the time the searches were run.
PubMed IDs for publications relevant to the gene.
For mouse genes, this information is hand-curated by MGI and uploaded to the NCBI Entrez Gene database; 906 of the genes (95%) had corresponding PubMed publications.
Human orthologues for the mouse gene: 866 of the genes (91%) had a known human orthologue.Footnote 18
MGI information: 883 of the genes (93%) had an MGI identifier.
Coordinates for the gene’s position in the genome; this information is used for visualizations of the mouse gene patent landscape – it is available for all matched genes.
We also calculated statistics for genes using the MGI Gene Ontology Slim Chart Tool. These statistics were in addition to information specific to each genetic sequence mapped to each gene: Strand matched; Direction of match; Position of matched sequence in the genome; Chromosome (1-Y, mitochondrial); and Quality of match (score).
Comparison Set of Non-patented Mouse Genes
For comparison purposes, Dr. Songyan Liu randomly selected a comparable number of unpatented genes. First, we randomly determined 2000 Ensembl Gene database entries for mouse genes. Of these, we removed 56 that were in the list of patented genes. Second, we searched for the remaining 1944 genes in MGI and identified 2012 hits. We removed 489 genes from this list if they did not have an official MGI symbol, 47 genes because they were in fact pseudogenes, and 96 genes because they were duplicates, including genes with multiple loci or Y chromosome genes that were a duplicate of X chromosome genes. In total, therefore, we selected 1397 genes for the control set to compare against our 952 patented genes out of a total of 32,480 possible genes (including mitochondrial genes) from NCBI Mouse Build 37.
As earlier, we extracted the following information in February 2009 on the genes from bioinformatics databases:
1069 genes (77%) had been investigated for targeting.
All genes in this control set had hits in all three components of the MGI Gene Ontology Slim Chart (i.e., none were “novel genes”).
144 genes (10%) had a corresponding OMIM ID; 133 (9.5%) had associated detailed disease identifiers.
266 genes (19%) had associated phenotype information.
1079 (77%) had at least one component of associated Gene Ontology information.
1211 (87%) had associated PubMed publications.
Mouse Gene Literature
We downloaded the full XML records for the MGI mouse gene associated PMIDs from PubMed, which resulted in 23,805 publications on patented mouse genes and 10,684 on non-patented mouse genes in December 2008. We then downloaded full records for literature that cited those publications from Thomson’s ISI database. In detail, we
Parsed XML PubMed records into an SQL database, using a Python script written by Dr. Strotmann, to extract (1) author names, affiliation; (2) article title, major MeSH codes; and (3) journal name, issue, year, number, pages.
Located and downloaded full corresponding records in the Thomson ISI database so that we could download all citing literature. We located 98% of PubMed records in ISI.
Statistical Analysis
The goal of our statistical analysis, performed by consulting biostatistician Dr. Shawn Morrison, was to determine if the citation and publication rates for publications on mouse genes (1) changed after patenting and (2) differed between publications on patented and unpatented mouse genes. We considered eight time periods: ±1, ±3, ±5, and ± 7 years before and after patenting. For patented genes, date “0” was the date the US patent was granted. For non-patented genes, date “0” was the median time from the original publication to the date of the search. This gave us a distribution of publications that had from 0 to at least 14 years of publication and citation data. Given the length of time from scientific publication to patent grant, the two data sets had similar distributions around the patent date and the median publication date.
We retained only those articles that had sufficient data to estimate all year intervals for analysis. For example, if an article had ±4 years of data, it was included in the ±3 years analysis, but not the ±5 analysis. Some genes had sufficient data for the pre-patenting period but not the post-patenting period (and vice versa), and therefore sample sizes vary for each period.
Data in the original data set was on a per article basis (citations per year and per article). We re-summarized this information on a per gene basis rather than a per article basis. For example, in a given year, if one article about gene ‘X’ was cited 10 times, and another article about gene ‘X’ was cited 5 times, then the result was a total of 15 citations for that gene in that year. This per gene data was used to calculate citation rates and was the basis for summary statistics and t-tests (described later).
We calculated the publication and citation rates per gene for the eight periods. Calculation of citation rate requires information regarding the change in the number of publications/citations from one year to the next. For example, the citation rate in the first year post-patenting would be the rate from Year 0 to Year 1, the rate for the second year would be the rate from Year 1 to Year 2, and so on. More formally, the citation rate was the natural log of the ratio between the years of interest – this provides an estimate of the instantaneous rate of change at that point in time (i.e., the slope).
Some genes had a number of publications/citations in a given year but declined to zero citations in the next. This created difficulties in calculating rates (i.e., division by zero), and these genes were excluded from analysis. Fortunately, this only applied to a relatively small number of genes. The exception to this filtering rule occurs when both the starting and ending years had zero citations. In this case, the rate was unchanged (and calculated as a rate of change = 0.00).
Therefore, the years used in the calculation of publication rate for this analysis are shown in Table 10.A1 (note that the same rate calculation was applied to citations).
Table 10.A1 Rate calculations for publications
| Period of Interest (relative to patent year) | Year of Citation Data per Gene | Rate Calculation | |
|---|---|---|---|
| From | To | ||
| –1 | 0 | –1 | ln(pubs in year-1/ pubs in year 0) |
| +1 | 0 | +1 | ln(pubs in year 0/ pubs in year+1) |
| –3 | –3 | –2 | ln(pubs in year-3/ pubs in year -2) |
| +3 | +2 | +3 | ln(pubs in year+2/ pubs in year+3) |
| –5 | –5 | –4 | ln(pubs in year-5/ pubs in year-4) |
| +5 | +4 | +5 | ln(pubs in year+4/ pubs in year+5) |
| –7 | –6 | –7 | ln(pubs in year-7/ pubs in year-6) |
| +7 | +6 | +7 | ln(pubs in year+6/ pubs in year+7) |
Sample Calculations and Conversions
If an article was cited 10 times in the year of patent grant (Year 0) and cited 11 times in the year following (Year 1), then the rate of citation during the first year post-patenting (Year 0 to Year 1) would be:
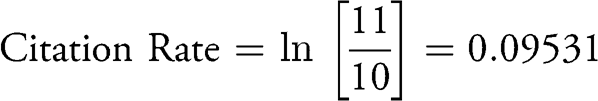
To estimate the percentage increase in citations over a given period, it is necessary to convert the instantaneous rate of change (r) to the finite rate of change (λ) as follows:
λ = er, where “λ” is the finite rate of change and “r” is the instantaneous rate of change. λ may be thought of as a “multiplier” between years. In the previous example, one would have to have an increase of 10% for the number of citations to increase from 10 to 11. The multiplier in this situation is 1.1, or a 10% increase.
For example, if r = 0.09531, then the finite citation rate is calculated as er = e°.09531 = 1.1 per year, which is interpreted as a 10% increase in the number of citations. To convert back, the equation is as follows: ln(λ) = r = ln(1.1) = 0.09531.
The relationship between r and λ is shown in the accompanying table.
| Citation Rate Is | r | λ |
|---|---|---|
| Decreasing | <0.0 | <1.0 |
| Stable | =0.0 | =1.0 |
| Increasing | >0.0 | >1.0 |
Thus, the citation rate is increasing when r >0 and/or λ >1.0.
Analysis and Results
Summary statistics for publication and citation rate per gene were calculated for each time period (Tables 10.A2–10.A5). Time periods were compared using Welch’s t-testsFootnote 19 which are similar to the common Student’s t-test but without the requirements for equal variances or equal sample sizes. A t-test was conducted for each period (±1, ±3, ±5 and ±7 years pre- and post-patenting) within publications and citations. Welch’s t-tests were then used to compare each time period between patented and unpatented genes for both publication and citation rates. To compensate for false positive significance as a result of large sample sizes and multiple t-tests, we increased the significant P-value from 0.05 to 0.01. In the following tables, significant differences are bolded. In addition for each time period, we compared publication and citation rates between patented and unpatented genes using Welch’s t-test (Table 10.A6).
Table 10.A2 Summary statistics for publication rate per patented gene
| Period Relative to Patent Grant Year | Mean (r) | Std. Error | # genes | # publications | Years Compared | P-value | t-value | df |
|---|---|---|---|---|---|---|---|---|
| 1 year prior | –0.009 | 0.020 | 633 | 2084 | –1 to +1 | 0.091 | 1.691 | 4159 |
| 1 year post | –0.059 | 0.021 | 606 | 2088 | ||||
| 3 years prior | –0.008 | 0.019 | 619 | 2332 | -3 to +3 | 0.007 | 2.710 | 4043 |
| 3 years post | –0.082 | 0.019 | 633 | 1767 | ||||
| 5 years prior | –0.034 | 0.021 | 649 | 2279 | –5 to +5 | 0.873 | 0.160 | 3504 |
| 5 years post | –0.030 | 0.014 | 696 | 1240 | ||||
| 7 years prior | 0.027 | 0.011 | 706 | 1890 | –7 to +7 | 0.564 | 0.578 | 2386 |
| 7 years post | –0.027 | 0.011 | 745 | 757 |
Table 10.A3 Summary statistics for citation rate per patented gene.
| Period Relative to Patent Grant Year | Mean (r) | Std. Error | # genes | # citations | Years Compared | P-value | t-value | df |
|---|---|---|---|---|---|---|---|---|
| 1 year prior | 0.417 | 0.028 | 680 | 118037 | –1 to +1 | 0.052 | 1.94 | 237635 |
| 1 year post | 0.344 | 0.024 | 738 | 144949 | ||||
| 3 years prior | 0.473 | 0.032 | 490 | 73537 | –3 to +3 | <.0001 | 7.276 | 157969 |
| 3 years post | 0.179 | 0.024 | 779 | 193711 | ||||
| 5 years prior | 0.568 | 0.043 | 325 | 39525 | –5 to +5 | <.0001 | 7.965 | 66863 |
| 5 years post | 0.179 | 0.024 | 740 | 222987 | ||||
| 7 years prior | 0.572 | 0.063 | 184 | 21173 | –7 to +7 | <.0001 | 8.806 | 28759 |
| 7 years post | –0.030 | 0.026 | 647 | 236608 |
Table 10.A4 Summary statistics for publication rate per unpatented gene.
| Period Relative to Median Publication Date | Mean (r) | Std. Error | # genes | # publications | Years Compared | P-value | t-value | df |
|---|---|---|---|---|---|---|---|---|
| 1 year prior | –0.026 | 0.020 | 556 | 1025 | –1 to +1 | 0.668 | 0.429 | 1988 |
| 1 year post | –0.037 | 0.015 | 798 | 1444 | ||||
| 3 years prior | –0.018 | 0.015 | 700 | 1432 | –3 to +3 | 0.770 | 0.292 | 2126 |
| 3 years post | –0.013 | 0.010 | 875 | 702 | ||||
| 5 years prior | 0.0003 | 0.012 | 912 | 1063 | –5 to +5 | 0.624 | 0.491 | 1280 |
| 5 years post | –0.006 | 0.005 | 1080 | 220 | ||||
| 7 years prior | 0.029 | 0.008 | 1050 | 894 | –7 to +7 | 0.038 | 2.086 | 261 |
| 7 years post | –0.001 | 0.004 | 1125 | 122 |
Table 10.A5 Summary statistics for citation rate per unpatented gene.
| Period Relative to Median Publication Date | Mean (r) | Std. Error | # genes | # citations | Years Compared | P-value | t-value | df |
|---|---|---|---|---|---|---|---|---|
| 1 year prior | 0.632 | 0.027 | 960 | 44835 | –1 to +1 | <.0001 | 15.801 | 98752 |
| 1 year post | 0.696 | 0.023 | 1027 | 73640 | ||||
| 3 years prior | 0.602 | 0.046 | 332 | 14329 | –3 to +3 | <.0001 | 11.593 | 17770 |
| 3 years post | 0.040 | 0.016 | 1187 | 171824 | ||||
| 5 years prior | 0.332 | 0.051 | 154 | 8652 | –5 to +5 | <.0001 | 4.047 | 10459 |
| 5 years post | 0.116 | 0.016 | 1142 | 224631 | ||||
| 7 years prior | 0.204 | 0.059 | 109 | 5287 | –7 to +7 | <.0001 | 16.046 | 7113 |
| 7 years post | –0.812 | 0.024 | 1119 | 235114 |
Table 10.A6 Summary statistics for comparison within each time period of publication rate for patented and unpatented genes
Statistically significant differences are bolded.
| Period Relative to Year 0* | P-value | t-value | df |
|---|---|---|---|
| 7 years prior | 0.896 | 0.130 | 2731 |
| 5 years prior | 0.147 | 1.451 | 3260 |
| 3 years prior | 0.679 | 0.414 | 3760 |
| 1 year prior | 0.560 | 0.584 | 2747 |
| 1 year post | 0.389 | 0.862 | 3418 |
| 3 years post | 0.001 | 3.211 | 2407 |
| 5 years post | 0.114 | 1.583 | 1449 |
| 7 years post | 0.023 | 2.284 | 873 |
Table 10.A7 Summary statistics for comparison within each time period of citation rate for patented and unpatented genes
Statistically significant differences are bolded.
| Period Relative to Year 0* | P-value | t-value | df |
|---|---|---|---|
| 7 years prior | <.0001 | 4.266 | 18451 |
| 5 years prior | 0.0004 | 3.547 | 22556 |
| 3 years prior | 0.022 | 2.294 | 30776 |
| 1 year prior | <.0001 | 5.467 | 133264 |
| 1 year post | <.0001 | 10.570 | 204122 |
| 3 years post | <.0001 | 4.851 | 325261 |
| 5 years post | 0.028 | 2.203 | 392287 |
| 7 years post | <.0001 | 22.378 | 467887 |
* Note that in Tables 10.A6 and 10.A7, year 0 is the year the patent was granted for patented genes and the median year for publications from first publication to date of search for unpatented genes (December 2008) for unpatented genes.



